ADP_B_15(5X17)
KC1D_MOUSE
[Raw transfer]

|
KJD_B_5(6NCG)
VRK2_HUMAN
[Raw transfer]

|
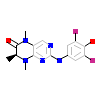
E5M_D_13(6BRU)
VRK1_HUMAN
[Raw transfer]
|
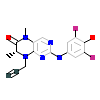
EA7_D_15(6CFM)
VRK1_HUMAN
[Raw transfer]
|
E5M_B_7(6BRU)
VRK1_HUMAN
[Raw transfer]

|
E8V_A_5(6BU6)
VRK1_HUMAN
[Raw transfer]

|
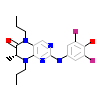
G6J_B_13(6DD4)
VRK1_HUMAN
[Raw transfer]
|
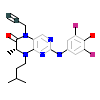
F87_B_10(6CNX)
VRK1_HUMAN
[Raw transfer]
|
F8Y_A_5(6CQH)
VRK1_HUMAN
[Raw transfer]

|
E8D_A_7(6BTW)
VRK1_HUMAN
[Raw transfer]
|
E8D_D_19(6BTW)
VRK1_HUMAN
[Raw transfer]

|
F7D_B_7(6CMM)
VRK1_HUMAN
[Raw transfer]

|
KJD_A_3(6NCG)
VRK2_HUMAN
[Raw transfer]

|
E8D_B_11(6BTW)
VRK1_HUMAN
[Raw transfer]

|
EA7_A_6(6CFM)
VRK1_HUMAN
[Raw transfer]

|
E1D_B_10(6BP0)
VRK1_HUMAN
[Raw transfer]

|
F87_A_5(6CNX)
VRK1_HUMAN
[Raw transfer]

|
G6J_D_25(6DD4)
VRK1_HUMAN
[Raw transfer]

|
E1D_D_20(6BP0)
VRK1_HUMAN
[Raw transfer]

|
7DZ_D_19(5UVF)
VRK1_HUMAN
[Raw transfer]
|
KWJ_A_5(6NPN)
VRK1_HUMAN
[Raw transfer]
|
7DZ_A_5(5UVF)
VRK1_HUMAN
[Raw transfer]

|
F8Y_B_10(6CQH)
VRK1_HUMAN
[Raw transfer]

|
7DZ_C_11(5UVF)
VRK1_HUMAN
[Raw transfer]

|
7DZ_A_2(5UU1)
VRK2_HUMAN
[Raw transfer]

|
KWJ_C_18(6NPN)
VRK1_HUMAN
[Raw transfer]

|
7DZ_A_2(5UU1)
VRK2_HUMAN
[Raw transfer]

|
7DZ_A_5(5UVF)
VRK1_HUMAN
[Raw transfer]

|
EA7_B_10(6CFM)
VRK1_HUMAN
[Raw transfer]

|
KWJ_D_25(6NPN)
VRK1_HUMAN
[Raw transfer]

|
KWJ_B_8(6NPN)
VRK1_HUMAN
[Raw transfer]

|
REB_B_8(3OP5)
VRK1_HUMAN
[Raw transfer]

|
8E1_A_5(5UKF)
VRK1_HUMAN
[Raw transfer]
|
REB_A_5(3OP5)
VRK1_HUMAN
[Raw transfer]

|
8E1_C_14(5UKF)
VRK1_HUMAN
[Raw transfer]

|
7DZ_B_9(5UVF)
VRK1_HUMAN
[Raw transfer]

|
REB_C_11(3OP5)
VRK1_HUMAN
[Raw transfer]

|
REB_D_15(3OP5)
VRK1_HUMAN
[Raw transfer]

|
1QO_B_8(4KB8)
KC1D_HUMAN
[Raw transfer]

|
ADP_B_15(5X17)
KC1D_MOUSE
[Raw transfer]

|
1QO_D_14(4KB8)
KC1D_HUMAN
[Raw transfer]

|
8E1_D_19(5UKF)
VRK1_HUMAN
[Raw transfer]

|
ANP_A_5(6AC9)
VRK1_HUMAN
[Raw transfer]

|
8E1_B_8(5UKF)
VRK1_HUMAN
[Raw transfer]

|
37J_A_9(4TWC)
KC1D_HUMAN
[Raw transfer]

|
386_A_12(4TW9)
KC1D_HUMAN
[Raw transfer]

|
37J_B_18(4TWC)
KC1D_HUMAN
[Raw transfer]

|
9WG_C_16(5W4W)
KC1D_HUMAN
[Raw transfer]

|
0CK_B_8(3UYT)
KC1D_HUMAN
[Raw transfer]

|
AUE_A_3(5IH5)
KC1D_HUMAN
[Raw transfer]

|
1QM_B_8(4KBA)
KC1D_HUMAN
[Raw transfer]

|
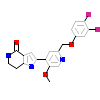
15G_B_4(4HGT)
KC1D_HUMAN
[Raw transfer]
|
1QG_C_14(4KBK)
KC1D_HUMAN
[Raw transfer]

|
GEW_B_5(6HMP)
KC1D_HUMAN
[Raw transfer]

|
AUG_A_6(5IH6)
KC1D_HUMAN
[Raw transfer]

|
GEW_A_3(6HMP)
KC1D_HUMAN
[Raw transfer]

|
1QN_C_11(4KB8)
KC1D_HUMAN
[Raw transfer]

|
1QN_A_5(4KB8)
KC1D_HUMAN
[Raw transfer]

|
1QG_A_5(4KBK)
KC1D_HUMAN
[Raw transfer]

|
15G_A_3(4HGT)
KC1D_HUMAN
[Raw transfer]

|
16W_B_4(4HNF)
KC1D_HUMAN
[Raw transfer]

|
1QM_C_11(4KBA)
KC1D_HUMAN
[Raw transfer]

|
16W_A_3(4HNF)
KC1D_HUMAN
[Raw transfer]

|
0CK_D_13(3UYT)
KC1D_HUMAN
[Raw transfer]

|
1QJ_B_7(4KBC)
KC1D_HUMAN
[Raw transfer]

|
16W_A_3(4HNI)
KC1E_HUMAN
[Raw transfer]

|
GE5_A_3(6HMR)
KC1D_HUMAN
[Raw transfer]

|
0CK_A_5(3UYT)
KC1D_HUMAN
[Raw transfer]

|
HSJ_A_13(4TW9)
KC1D_HUMAN
[Raw transfer]

|