7AV_A_3(5TF9)
WNK1_HUMAN
[Raw transfer]

|
A7Y_B_8(5WE8)
WNK1_HUMAN
[Raw transfer]

|
7AV_B_7(5TF9)
WNK1_HUMAN
[Raw transfer]

|
GOL_A_5(5O2C)
WNK3_HUMAN
[Raw transfer]

|
GOL_A_5(5O2C)
WNK3_HUMAN
[Raw transfer]

|
GOL_B_8(6CN9)
WNK1_RAT
[Raw transfer]

|
GOL_B_10(6CN9)
WNK1_RAT
[Raw transfer]

|
9HE_B_5(5O2B)
WNK3_HUMAN
[Raw transfer]
|
GOL_A_3(6CN9)
WNK1_RAT
[Raw transfer]

|
A7Y_A_5(5WE8)
WNK1_HUMAN
[Raw transfer]

|
A6S_A_4(5WDY)
WNK1_HUMAN
[Raw transfer]

|
EU4_A_3(6CAD)
BRAF_HUMAN
[Raw transfer]

|
L1E_B_3(3IDP)
[Raw transfer]

|
3K3_A_3(4R5Y)
BRAF_HUMAN
[Raw transfer]

|
L1E_A_4(3IDP)
BRAF_HUMAN
[Raw transfer]

|
92D_A_3(5VAL)
BRAF_HUMAN
[Raw transfer]

|
A6S_B_9(5WDY)
WNK1_HUMAN
[Raw transfer]

|
KZP_B_3(6NSQ)
[Raw transfer]

|
BAX_A_3(1UWH)
BRAF_HUMAN
[Raw transfer]

|
5FJ_A_2(5DRB)
WNK1_RAT
[Raw transfer]
|
5FJ_A_2(5DRB)
WNK1_RAT
[Raw transfer]

|
9HE_A_3(5O2B)
WNK3_HUMAN
[Raw transfer]

|
1SU_A_3(4KSP)
BRAF_HUMAN
[Raw transfer]

|
3K3_B_4(4R5Y)
[Raw transfer]

|
B1E_B_5(5HID)
[Raw transfer]

|
K81_A_3(6N0P)
BRAF_HUMAN
[Raw transfer]

|
0NF_A_3(3Q96)
BRAF_HUMAN
[Raw transfer]

|
B1E_B_4(4G9R)
[Raw transfer]

|
B1E_B_4(4RZW)
[Raw transfer]

|
92D_B_4(5VAL)
[Raw transfer]

|
BAX_A_2(5HI2)
[Raw transfer]

|
B96_C_7(4JVG)
BRAF_HUMAN
[Raw transfer]
|
0T2_A_3(4FC0)
BRAF_HUMAN
[Raw transfer]

|
B96_D_8(4JVG)
[Raw transfer]

|
0WP_B_4(4G9C)
[Raw transfer]

|
1SW_A_3(4KSQ)
BRAF_HUMAN
[Raw transfer]

|
B1E_A_3(4G9R)
BRAF_HUMAN
[Raw transfer]

|
4Z5_A_3(5C9C)
BRAF_HUMAN
[Raw transfer]

|
B96_B_5(4JVG)
BRAF_HUMAN
[Raw transfer]

|
4Z5_B_4(5C9C)
[Raw transfer]

|
B1E_A_3(5HID)
BRAF_HUMAN
[Raw transfer]

|
B96_A_6(4JVG)
BRAF_HUMAN
[Raw transfer]

|
1SU_B_4(4KSP)
[Raw transfer]

|
0T2_B_4(4FC0)
[Raw transfer]

|
55J_A_3(5CT7)
BRAF_HUMAN
[Raw transfer]

|
K7S_A_3(6N0Q)
?
[Raw transfer]

|
0NF_B_4(3Q96)
[Raw transfer]

|
B1E_A_3(4RZW)
BRAF_HUMAN
[Raw transfer]

|
831_B_6(3II5)
[Raw transfer]
|
3RJ_C_9(4WNP)
ULK1_HUMAN
[Raw transfer]
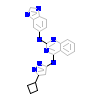
|
FP4_B_4(3PRI)
[Raw transfer]
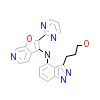
|
K7S_B_4(6N0Q)
?
[Raw transfer]

|
BAX_B_4(1UWH)
[Raw transfer]

|
1SW_B_4(4KSQ)
[Raw transfer]

|
T1Q_A_3(4E4X)
BRAF_HUMAN
[Raw transfer]

|
BAX_B_4(1UWJ)
[Raw transfer]

|
RI8_B_4(4EHE)
[Raw transfer]

|
324_B_4(4WO5)
[Raw transfer]

|
0WP_A_3(4G9C)
BRAF_HUMAN
[Raw transfer]

|
RI8_A_3(4EHE)
BRAF_HUMAN
[Raw transfer]

|
6DC_A_3(5ITA)
BRAF_HUMAN
[Raw transfer]

|
BR2_B_4(3SKC)
[Raw transfer]

|
2VX_A_3(4PP7)
BRAF_HUMAN
[Raw transfer]

|
K81_B_4(6N0P)
[Raw transfer]

|
BAX_A_3(1UWJ)
BRAF_HUMAN
[Raw transfer]

|
8EN_A_3(6B8U)
BRAF_HUMAN
[Raw transfer]

|
032_A_3(3OG7)
BRAF_HUMAN
[Raw transfer]
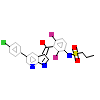
|
324_A_3(3C4C)
BRAF_HUMAN
[Raw transfer]

|
ANP_A_3(5O26)
WNK3_HUMAN
[Raw transfer]

|
P06_A_3(4XV2)
BRAF_HUMAN
[Raw transfer]

|
T1Q_B_4(4E4X)
[Raw transfer]

|
325_A_3(4FK3)
BRAF_HUMAN
[Raw transfer]

|
ANP_B_7(5O26)
WNK3_HUMAN
[Raw transfer]

|
8EN_B_4(6B8U)
[Raw transfer]

|
2VX_B_4(4PP7)
[Raw transfer]

|
P06_B_4(5CSW)
[Raw transfer]

|
324_A_3(4WO5)
BRAF_HUMAN
[Raw transfer]

|
ANP_A_4(5WE8)
WNK1_HUMAN
[Raw transfer]

|
CIT_A_6(6QAS)
ULK1_HUMAN
[Raw transfer]
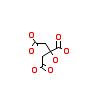
|
1OO_A_2(4XV9)
[Raw transfer]
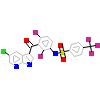
|
P06_B_4(4XV2)
[Raw transfer]

|
92J_A_3(5VAM)
BRAF_HUMAN
[Raw transfer]

|
DFS_B_4(4MBJ)
[Raw transfer]

|
CIT_B_24(6QAS)
ULK1_HUMAN
[Raw transfer]

|
ANP_B_4(5W7T)
WNK1_RAT
[Raw transfer]

|
ANP_B_8(5WDY)
WNK1_HUMAN
[Raw transfer]

|
0JA_B_4(4DBN)
[Raw transfer]

|
ANP_B_8(5TF9)
WNK1_HUMAN
[Raw transfer]

|
ANP_A_4(5TF9)
WNK1_HUMAN
[Raw transfer]

|
ANP_B_7(5WE8)
WNK1_HUMAN
[Raw transfer]

|
ANP_A_3(5W7T)
WNK1_RAT
[Raw transfer]

|
ANP_A_3(5W7T)
WNK1_RAT
[Raw transfer]

|
EDO_B_17(5O1V)
WNK3_HUMAN
[Raw transfer]

|
CQE_A_3(4CQE)
BRAF_HUMAN
[Raw transfer]

|
ANP_A_3(5WDY)
WNK1_HUMAN
[Raw transfer]

|
SM5_A_3(3OMV)
RAF1_HUMAN
[Raw transfer]

|
EDO_A_6(5O1V)
WNK3_HUMAN
[Raw transfer]

|
EDO_A_6(5O1V)
WNK3_HUMAN
[Raw transfer]

|
EDO_A_16(5O23)
WNK3_HUMAN
[Raw transfer]

|
ADP_A_2(2WQN)
NEK7_HUMAN
[Raw transfer]

|
EDO_B_15(5O21)
WNK3_HUMAN
[Raw transfer]

|
EDO_B_26(5O23)
WNK3_HUMAN
[Raw transfer]

|
DMS_A_4(5JRQ)
BRAF_HUMAN
[Raw transfer]
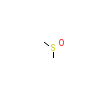
|
GLC_A_3(5CSX)
[Raw transfer]
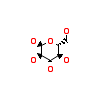
|
DMS_A_6(5JSM)
BRAF_HUMAN
[Raw transfer]
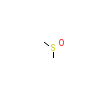
|
DMS_D_20(5JSM)
[Raw transfer]
|
DMS_B_11(5JSM)
BRAF_HUMAN
[Raw transfer]
|