P01_A_2(2IZU)
KC1G3_HUMAN
[Raw transfer]
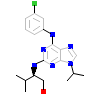
|
0VM_A_2(4G16)
KC1G3_HUMAN
[Raw transfer]
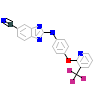
|
BRK_A_2(2IZR)
KC1G3_HUMAN
[Raw transfer]
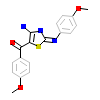
|
BRK_A_2(2IZR)
KC1G3_HUMAN
[Raw transfer]

|
23D_A_2(2IZT)
KC1G3_HUMAN
[Raw transfer]
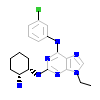
|
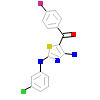
BRQ_A_2(2IZS)
KC1G3_HUMAN
[Raw transfer]
|
F92_A_7(6GRO)
KC1G3_HUMAN
[Raw transfer]

|
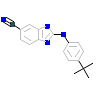
0VN_A_2(4G17)
KC1G3_HUMAN
[Raw transfer]
|
0YO_A_2(4HGL)
KC1G3_HUMAN
[Raw transfer]

|
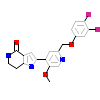
15G_A_2(4HGS)
KC1G3_HUMAN
[Raw transfer]
|
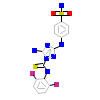
DKI_A_2(2CHL)
KC1G3_HUMAN
[Raw transfer]
|
16W_A_3(4HNI)
KC1E_HUMAN
[Raw transfer]

|