3JZ_A_2(3FZR)
[Raw transfer]
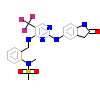
|
AGS_A_2(3FZP)
[Raw transfer]
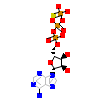
|
349_A_2(3ET7)
[Raw transfer]

|
YAM_A_2(5TOB)
[Raw transfer]

|
7FM_A_2(5TO8)
[Raw transfer]

|
4JZ_A_2(3FZT)
[Raw transfer]
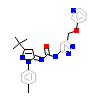
|
0YJ_A_2(4H1M)
[Raw transfer]
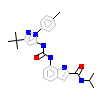
|
0YH_A_2(4H1J)
[Raw transfer]

|
B96_A_2(3FZS)
[Raw transfer]
|