L1G_A_3(2C0I)
HCK_HUMAN
[Raw transfer]
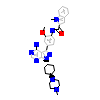
|
L2G_A_3(2C0O)
HCK_HUMAN
[Raw transfer]

|
VRZ_B_8(3VRZ)
HCK_HUMAN
[Raw transfer]
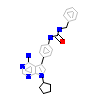
|
OOU_A_2(5H0G)
HCK_HUMAN
[Raw transfer]
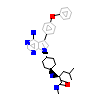
|
L3G_A_3(2C0T)
HCK_HUMAN
[Raw transfer]
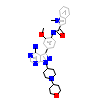
|
OOV_A_2(5H0H)
HCK_HUMAN
[Raw transfer]
|
VRZ_A_5(3VRZ)
HCK_HUMAN
[Raw transfer]

|
L3G_B_6(2C0T)
HCK_HUMAN
[Raw transfer]

|
L1G_B_5(2C0I)
HCK_HUMAN
[Raw transfer]

|
L2G_B_5(2C0O)
HCK_HUMAN
[Raw transfer]

|
VSF_B_8(3VS4)
HCK_HUMAN
[Raw transfer]
|
VSE_B_10(4LUE)
HCK_HUMAN
[Raw transfer]

|
OOO_A_2(5H09)
HCK_HUMAN
[Raw transfer]
|
VSH_A_6(3VS6)
HCK_HUMAN
[Raw transfer]
|
VSE_A_3(5ZJ6)
HCK_HUMAN
[Raw transfer]

|
VSE_B_4(5ZJ6)
HCK_HUMAN
[Raw transfer]

|
VSB_A_3(3VS2)
HCK_HUMAN
[Raw transfer]

|
VSF_A_5(3VS4)
HCK_HUMAN
[Raw transfer]

|
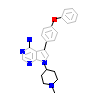
VSG_A_3(3VS5)
HCK_HUMAN
[Raw transfer]
|
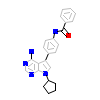
VS0_A_5(3VS0)
HCK_HUMAN
[Raw transfer]
|
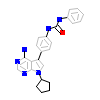
VSA_A_5(3VS1)
HCK_HUMAN
[Raw transfer]
|
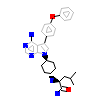
OOS_A_2(5H0E)
HCK_HUMAN
[Raw transfer]
|
VSA_B_8(3VS1)
HCK_HUMAN
[Raw transfer]

|
OOQ_A_2(5H0B)
HCK_HUMAN
[Raw transfer]

|
VSE_A_3(3VS3)
HCK_HUMAN
[Raw transfer]

|
VSH_B_10(3VS6)
HCK_HUMAN
[Raw transfer]

|
VSB_B_7(3VS2)
HCK_HUMAN
[Raw transfer]

|
VS0_B_8(3VS0)
HCK_HUMAN
[Raw transfer]

|
B43_A_3(3VRY)
HCK_HUMAN
[Raw transfer]
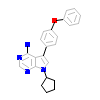
|
B43_B_7(3VRY)
HCK_HUMAN
[Raw transfer]

|
VSE_B_7(3VS3)
HCK_HUMAN
[Raw transfer]

|
VSG_B_6(3VS5)
HCK_HUMAN
[Raw transfer]

|
VSE_A_4(4LUE)
HCK_HUMAN
[Raw transfer]

|
ANP_B_8(1AD5)
HCK_HUMAN
[Raw transfer]

|
PP1_B_2(1QCF)
HCK_HUMAN
[Raw transfer]

|
ANP_A_7(1AD5)
HCK_HUMAN
[Raw transfer]

|
KS1_B_6(3VS7)
HCK_HUMAN
[Raw transfer]
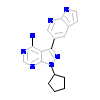
|
SK8_B_8(4LUD)
HCK_HUMAN
[Raw transfer]

|
SK8_A_5(4LUD)
HCK_HUMAN
[Raw transfer]

|
1BM_A_2(2HK5)
HCK_HUMAN
[Raw transfer]
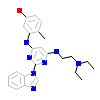
|
QUE_B_6(2HCK)
HCK_HUMAN
[Raw transfer]

|
QUE_A_5(2HCK)
HCK_HUMAN
[Raw transfer]

|