EBI_A_2(3I81)
[Raw transfer]
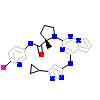
|
LGV_A_2(3NW7)
[Raw transfer]
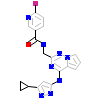
|
LGX_A_2(3NW5)
[Raw transfer]

|
01P_D_8(3QQU)
[Raw transfer]
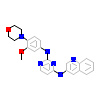
|
LGW_A_2(3NW6)
[Raw transfer]

|
01P_A_5(3QQU)
IGF1R_HUMAN
[Raw transfer]

|
01P_B_6(3QQU)
IGF1R_HUMAN
[Raw transfer]

|
01P_C_7(3QQU)
IGF1R_HUMAN
[Raw transfer]

|
BMI_B_2(2OJ9)
[Raw transfer]

|
OZN_A_5(5FXS)
[Raw transfer]
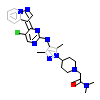
|
8LN_A_2(5FXR)
[Raw transfer]

|
ACP_C_3(1K3A)
[Raw transfer]

|
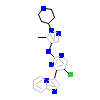
GD5_A_2(5FXQ)
[Raw transfer]
|
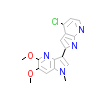
PDR_A_5(3LVP)
IGF1R_HUMAN
[Raw transfer]
|
ANP_B_9(1JQH)
IGF1R_HUMAN
[Raw transfer]

|
DYK_A_3(4D2R)
[Raw transfer]

|
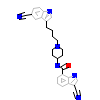
CCX_B_7(3LW0)
IGF1R_HUMAN
[Raw transfer]
|
CCX_D_13(3LW0)
[Raw transfer]

|
CCX_C_10(3LW0)
IGF1R_HUMAN
[Raw transfer]

|
CCX_A_5(3LW0)
IGF1R_HUMAN
[Raw transfer]

|