ADP_B_15(5X17)
KC1D_MOUSE
[Raw transfer]

|
PFO_A_3(4TN6)
KC1D_HUMAN
[Raw transfer]

|
1QM_B_8(4KBA)
KC1D_HUMAN
[Raw transfer]

|
0CK_B_8(3UYT)
KC1D_HUMAN
[Raw transfer]

|
0CK_A_5(3UYT)
KC1D_HUMAN
[Raw transfer]

|
1QM_C_11(4KBA)
KC1D_HUMAN
[Raw transfer]

|
9WG_B_9(5W4W)
KC1D_HUMAN
[Raw transfer]

|
0CK_D_13(3UYT)
KC1D_HUMAN
[Raw transfer]

|
1QN_A_5(4KB8)
KC1D_HUMAN
[Raw transfer]

|
0CK_C_10(3UYT)
KC1D_HUMAN
[Raw transfer]

|
1QM_A_5(4KBA)
KC1D_HUMAN
[Raw transfer]

|
1QG_B_10(4KBK)
KC1D_HUMAN
[Raw transfer]

|
9WG_A_5(5W4W)
KC1D_HUMAN
[Raw transfer]

|
0CK_B_4(3UZP)
KC1D_HUMAN
[Raw transfer]

|
0CK_A_3(3UZP)
KC1D_HUMAN
[Raw transfer]

|
1QM_D_14(4KBA)
KC1D_HUMAN
[Raw transfer]

|
1QJ_A_3(4KBC)
KC1D_HUMAN
[Raw transfer]

|
1QG_A_5(4KBK)
KC1D_HUMAN
[Raw transfer]

|
9WG_C_16(5W4W)
KC1D_HUMAN
[Raw transfer]

|
1QN_C_11(4KB8)
KC1D_HUMAN
[Raw transfer]

|
C9Z_B_23(6F26)
?
[Raw transfer]

|
C9Z_A_16(6F26)
?
[Raw transfer]

|
CG5_B_19(6F1W)
?
[Raw transfer]
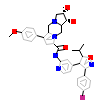
|
1QO_B_8(4KB8)
KC1D_HUMAN
[Raw transfer]

|
ADP_B_15(5X17)
KC1D_MOUSE
[Raw transfer]

|
1QG_C_14(4KBK)
KC1D_HUMAN
[Raw transfer]

|
1QO_D_14(4KB8)
KC1D_HUMAN
[Raw transfer]

|
1QG_D_18(4KBK)
KC1D_HUMAN
[Raw transfer]

|
9WG_D_21(5W4W)
KC1D_HUMAN
[Raw transfer]

|
386_A_12(4TW9)
KC1D_HUMAN
[Raw transfer]

|
ADP_A_9(5X17)
KC1D_MOUSE
[Raw transfer]

|
16W_B_9(4HNI)
KC1E_HUMAN
[Raw transfer]

|
1QJ_B_7(4KBC)
KC1D_HUMAN
[Raw transfer]

|
37J_B_18(4TWC)
KC1D_HUMAN
[Raw transfer]

|
15G_A_3(4HGT)
KC1D_HUMAN
[Raw transfer]
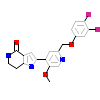
|
16W_A_3(4HNI)
KC1E_HUMAN
[Raw transfer]

|
37J_A_9(4TWC)
KC1D_HUMAN
[Raw transfer]

|
15G_B_4(4HGT)
KC1D_HUMAN
[Raw transfer]

|
386_B_20(4TW9)
KC1D_HUMAN
[Raw transfer]

|
AUG_A_6(5IH6)
KC1D_HUMAN
[Raw transfer]

|
16W_B_4(4HNF)
KC1D_HUMAN
[Raw transfer]

|
16W_A_3(4HNF)
KC1D_HUMAN
[Raw transfer]

|
CG5_B_19(6F1W)
?
[Raw transfer]

|