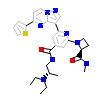
9EJ_B_5(5VLO)
KCC2D_HUMAN
[Raw transfer]
|
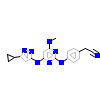
5CP_B_6(3BHH)
KCC2B_HUMAN
[Raw transfer]
|
9EJ_A_3(5VLO)
KCC2D_HUMAN
[Raw transfer]

|
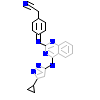
GVD_A_7(2VN9)
KCC2D_HUMAN
[Raw transfer]
|
GVD_B_8(2VN9)
KCC2D_HUMAN
[Raw transfer]

|
5CP_A_5(3BHH)
KCC2B_HUMAN
[Raw transfer]

|
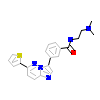
C2V_B_6(6AYW)
KCC2D_HUMAN
[Raw transfer]
|
D0S_B_7(6BAB)
KCC2D_MOUSE
[Raw transfer]

|
D0S_C_9(6BAB)
KCC2D_MOUSE
[Raw transfer]

|
C2V_A_3(6AYW)
KCC2D_HUMAN
[Raw transfer]

|
D0S_A_5(6BAB)
KCC2D_MOUSE
[Raw transfer]

|
DRN_A_2(2V7O)
KCC2G_HUMAN
[Raw transfer]

|