F82_B_4(6GQJ)
?
[Raw transfer]

|
F82_A_3(6GQJ)
?
[Raw transfer]

|
F88_B_4(6GQK)
?
[Raw transfer]

|
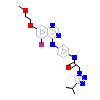
F8H_B_6(6GQM)
?
[Raw transfer]
|
F8H_A_3(6GQM)
[Raw transfer]

|
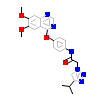
F8B_B_4(6GQL)
KIT_HUMAN
[Raw transfer]
|
F88_A_3(6GQK)
?
[Raw transfer]

|
F8B_A_3(6GQL)
KIT_HUMAN
[Raw transfer]

|
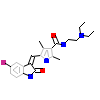
B49_A_2(3G0E)
?
[Raw transfer]
|