6V4_A_2(5GJF)
M3K7_HUMAN
[Raw transfer]
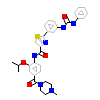
|
6KD_A_2(5JGD)
M3K7_HUMAN
[Raw transfer]
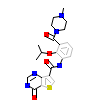
|
6V3_A_2(5GJD)
M3K7_HUMAN
[Raw transfer]
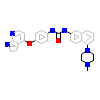
|
6KC_A_2(5JGA)
M3K7_HUMAN
[Raw transfer]
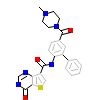
|
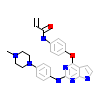
6HF_A_2(5J9L)
M3K7_HUMAN
[Raw transfer]
|
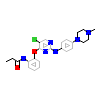
6H4_A_2(5J8I)
M3K7_HUMAN
[Raw transfer]
|
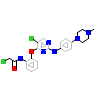
5KW_A_2(5E7R)
M3K7_HUMAN
[Raw transfer]
|
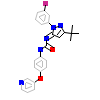
YIY_A_2(2YIY)
M3K7_HUMAN
[Raw transfer]
|
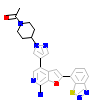
1UL_A_2(4L52)
M3K7_HUMAN
[Raw transfer]
|
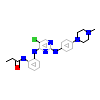
6H3_A_2(5J7S)
M3K7_HUMAN
[Raw transfer]
|
EDH_A_2(5V5N)
M3K7_HUMAN
[Raw transfer]

|
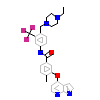
NG2_A_2(4O91)
M3K7_HUMAN
[Raw transfer]
|
1UH_A_2(4L3P)
M3K7_HUMAN
[Raw transfer]

|
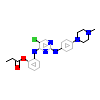
T92_A_2(5JH6)
M3K7_HUMAN
[Raw transfer]
|
6JV_A_2(5JGB)
M3K7_HUMAN
[Raw transfer]

|
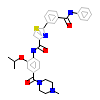
6V5_A_2(5GJG)
M3K7_HUMAN
[Raw transfer]
|
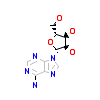
ADN_A_2(2EVA)
M3K7_HUMAN
[Raw transfer]
|
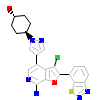
1UO_A_2(4L53)
M3K7_HUMAN
[Raw transfer]
|