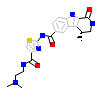
B18_A_5(2PZY)
MAPK2_HUMAN
[Raw transfer]
|
P4O_A_3(2JBO)
MAPK2_HUMAN
[Raw transfer]

|
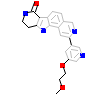
HGF_A_3(3M42)
MAPK2_HUMAN
[Raw transfer]
|
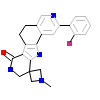
L8I_A_2(3M2W)
MAPK2_HUMAN
[Raw transfer]
|
P4O_A_4(3R2Y)
MAPK2_HUMAN
[Raw transfer]

|
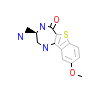
B98_X_2(3FYK)
MAPK2_HUMAN
[Raw transfer]
|
STU_C_7(2PZY)
MAPK2_HUMAN
[Raw transfer]

|
LX9_A_3(3KGA)
MAPK2_HUMAN
[Raw transfer]
|
STU_A_7(1NXK)
MAPK2_HUMAN
[Raw transfer]

|
B18_B_6(2PZY)
MAPK2_HUMAN
[Raw transfer]

|
B97_X_2(3FYJ)
MAPK2_HUMAN
[Raw transfer]

|
ADP_B_2(1NY3)
MAPK2_HUMAN
[Raw transfer]

|
F10_X_2(2P3G)
MAPK2_HUMAN
[Raw transfer]

|
MK3_A_2(3KA0)
MAPK2_HUMAN
[Raw transfer]
|