KRW_A_2(3ZXZ)
[Raw transfer]
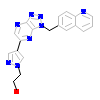
|
W9Z_A_2(3ZC5)
[Raw transfer]
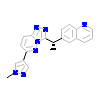
|
0JL_A_2(4DEI)
[Raw transfer]
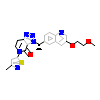
|
0JK_A_2(4DEH)
[Raw transfer]
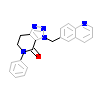
|
6XE_A_2(3ZBX)
[Raw transfer]
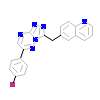
|
44X_A_2(4XYF)
[Raw transfer]
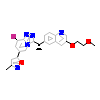
|
4K0_A_2(4AOI)
[Raw transfer]
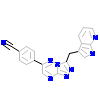
|
SX8_A_3(3DKG)
[Raw transfer]
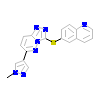
|
M61_A_2(3R7O)
[Raw transfer]
|
3QT_B_5(3QTI)
[Raw transfer]
|
5T1_A_2(5EYD)
[Raw transfer]
|
3E8_A_2(4R1V)
[Raw transfer]
|
3QT_A_3(3QTI)
MET_HUMAN
[Raw transfer]

|
L5G_A_2(3CD8)
[Raw transfer]
|
SX8_A_3(3DKF)
[Raw transfer]

|
5B4_A_3(5DG5)
MET_HUMAN
[Raw transfer]
|
46G_A_2(4XMO)
[Raw transfer]
|
DWF_A_2(4MXC)
[Raw transfer]
|
L1X_A_2(4EEV)
[Raw transfer]
|
B2D_A_2(3I5N)
[Raw transfer]
|
88Z_A_2(3LQ8)
[Raw transfer]

|
0JJ_A_2(4DEG)
[Raw transfer]
|
5SZ_A_2(5EYC)
[Raw transfer]
|
1RU_C_10(4KNB)
MET_HUMAN
[Raw transfer]
|
044_A_2(3U6I)
[Raw transfer]
|
0J3_A_2(4GG5)
[Raw transfer]
|
ATP_A_4(3DKC)
[Raw transfer]

|
ZZY_A_2(2WD1)
[Raw transfer]
|
319_A_2(3CTH)
[Raw transfer]
|
1RU_B_8(4KNB)
MET_HUMAN
[Raw transfer]

|
5TF_A_2(3ZCL)
[Raw transfer]
|
0J8_A_2(4GG7)
[Raw transfer]
|
5B4_B_4(5DG5)
[Raw transfer]

|
03X_A_2(3U6H)
[Raw transfer]
|
F47_A_2(4AP7)
[Raw transfer]
|
62E_A_3(5HLW)
[Raw transfer]
|
MT3_B_4(3EFJ)
[Raw transfer]
|
Q6W_A_2(3Q6W)
[Raw transfer]
|
MT4_B_4(3EFK)
[Raw transfer]
|
75H_A_2(5T3Q)
?
[Raw transfer]
|
DF6_A_2(3VW8)
[Raw transfer]
|
DFQ_A_5(3A4P)
[Raw transfer]
|
MT4_A_3(3EFK)
MET_HUMAN
[Raw transfer]

|
66L_A_2(5HTI)
[Raw transfer]
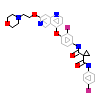
|
LKG_A_2(3CCN)
[Raw transfer]
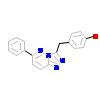
|
AM8_A_2(2RFS)
[Raw transfer]
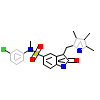
|
AM7_B_4(2RFN)
[Raw transfer]
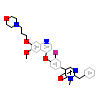
|
3EH_A_2(4R1Y)
[Raw transfer]
|
63B_Y_4(5HNI)
[Raw transfer]
|
MT3_A_3(3EFJ)
MET_HUMAN
[Raw transfer]

|
L8V_A_2(3L8V)
[Raw transfer]
|
353_A_2(3F82)
[Raw transfer]
|
CKK_A_2(3C1X)
[Raw transfer]
|
1FN_A_2(3CE3)
[Raw transfer]
|
63B_X_3(5HNI)
MET_HUMAN
[Raw transfer]

|
VGH_A_2(2WGJ)
[Raw transfer]
|
1RU_A_5(4KNB)
MET_HUMAN
[Raw transfer]

|
AM7_A_3(2RFN)
MET_HUMAN
[Raw transfer]

|
6XP_A_2(3ZZE)
[Raw transfer]
|
6TD_A_2(5YA5)
[Raw transfer]
|
PFY_A_2(2WKM)
[Raw transfer]
|
63K_A_2(5HOR)
[Raw transfer]
|
320_A_2(3CTJ)
[Raw transfer]
|
1RU_D_11(4KNB)
[Raw transfer]

|
KSA_B_2(1R0P)
[Raw transfer]
|
IHX_B_7(3F66)
[Raw transfer]
|
M97_B_4(3RHK)
[Raw transfer]
|
M97_A_3(3RHK)
MET_HUMAN
[Raw transfer]

|
IHX_A_3(3F66)
MET_HUMAN
[Raw transfer]

|
1JC_A_2(4IWD)
[Raw transfer]
|