X4Z_A_4(2X4Z)
[Raw transfer]
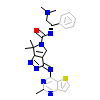
|
NJD_A_2(4NJD)
PAK4_HUMAN
[Raw transfer]
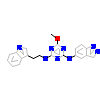
|
8FX_A_2(5XVG)
PAK4_HUMAN
[Raw transfer]
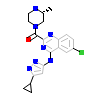
|
8FU_A_2(5XVA)
PAK4_HUMAN
[Raw transfer]
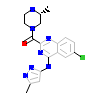
|
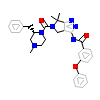
N53_A_2(4APP)
[Raw transfer]
|
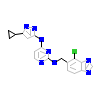
4T6_A_2(5BMS)
[Raw transfer]
|
ANP_A_5(4FIF)
PAK4_HUMAN
[Raw transfer]

|
ANP_B_6(4FIF)
PAK4_HUMAN
[Raw transfer]

|
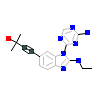
2OO_A_2(4O0Y)
[Raw transfer]
|
M77_A_2(5VEF)
?
[Raw transfer]

|
ANP_A_3(4FIG)
?
[Raw transfer]

|
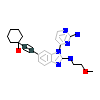
2OL_A_2(4O0V)
[Raw transfer]
|
ANP_A_3(4JDI)
PAK4_HUMAN
[Raw transfer]

|
STU_A_2(5VED)
PAK4_HUMAN
[Raw transfer]

|
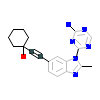
2OQ_A_2(4O0X)
?
[Raw transfer]
|
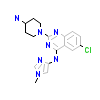
8FR_A_2(5XVF)
PAK4_HUMAN
[Raw transfer]
|
ANP_B_6(4FIG)
?
[Raw transfer]

|
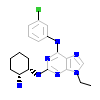
23D_A_2(2CDZ)
PAK4_HUMAN
[Raw transfer]
|
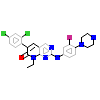
981_A_2(5VEE)
PAK4_HUMAN
[Raw transfer]
|
67U_A_2(5I0B)
PAK4_HUMAN
[Raw transfer]
|
ANP_B_4(5UPK)
?
[Raw transfer]

|
EDO_A_5(2J0I)
PAK4_HUMAN
[Raw transfer]

|
EDO_A_9(2Q0N)
PAK4_HUMAN
[Raw transfer]

|