ADN_A_2(4CKJ)
[Raw transfer]
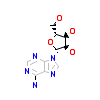
|
ADN_A_10(4CKI)
[Raw transfer]

|
PP1_A_2(5FM3)
[Raw transfer]

|
PP1_A_3(2IVV)
[Raw transfer]

|
X2L_A_6(2X2L)
[Raw transfer]
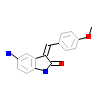
|
X2M_A_5(2X2M)
RET_HUMAN
[Raw transfer]

|
ACK_A_6(2IVS)
RET_HUMAN
[Raw transfer]
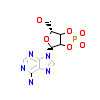
|
PP1_A_2(5FM2)
[Raw transfer]

|
ADN_A_5(6FEK)
[Raw transfer]

|
DTQ_A_2(5AMN)
[Raw transfer]

|
ZD6_A_4(2IVU)
[Raw transfer]

|