6U2_B_4(5KKT)
ROCK1_HUMAN
[Raw transfer]

|
6U2_A_3(5KKT)
ROCK1_HUMAN
[Raw transfer]

|
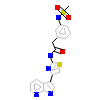
6U1_B_4(5KKS)
ROCK1_HUMAN
[Raw transfer]
|
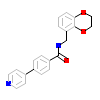
J0P_A_3(6E9W)
ROCK1_HUMAN
[Raw transfer]
|
6U1_A_3(5KKS)
ROCK1_HUMAN
[Raw transfer]

|
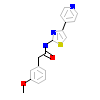
4KK_A_3(4YVE)
ROCK1_HUMAN
[Raw transfer]
|
8UV_A_3(5UZJ)
ROCK1_HUMAN
[Raw transfer]

|
4KK_B_4(4YVE)
ROCK1_HUMAN
[Raw transfer]

|
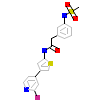
4TW_B_4(5BML)
ROCK1_HUMAN
[Raw transfer]
|
8UV_B_4(5UZJ)
ROCK1_HUMAN
[Raw transfer]

|
65R_A_3(5HVU)
ROCK1_HUMAN
[Raw transfer]

|
65R_B_4(5HVU)
ROCK1_HUMAN
[Raw transfer]

|
4KH_B_4(4YVC)
ROCK1_HUMAN
[Raw transfer]
|
4TW_A_3(5BML)
ROCK1_HUMAN
[Raw transfer]

|
4KH_A_3(4YVC)
ROCK1_HUMAN
[Raw transfer]

|
M77_D_4(2ESM)
ROCK1_HUMAN
[Raw transfer]

|
M77_C_3(2ESM)
ROCK1_HUMAN
[Raw transfer]

|
H52_B_4(3D9V)
ROCK1_HUMAN
[Raw transfer]

|
HFS_D_4(2ETK)
ROCK1_HUMAN
[Raw transfer]

|
H52_A_3(3D9V)
ROCK1_HUMAN
[Raw transfer]

|
ANP_A_5(2V55)
ROCK1_HUMAN
[Raw transfer]

|
Y27_C_3(2ETR)
ROCK1_HUMAN
[Raw transfer]

|
HFS_C_3(2ETK)
ROCK1_HUMAN
[Raw transfer]

|
Y27_D_4(2ETR)
ROCK1_HUMAN
[Raw transfer]

|