ACE_A_4(1HCS)
[Raw transfer]
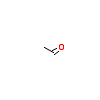
|
ACE_D_6(1A1C)
[Raw transfer]

|
787_B_2(1O4R)
[Raw transfer]
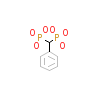
|
UR2_A_2(1SKJ)
[Raw transfer]
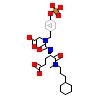
|
852_B_2(1O44)
[Raw transfer]
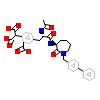
|
791_B_2(1O4P)
[Raw transfer]
|
876_A_2(1O4B)
[Raw transfer]
|
843_B_2(1O42)
[Raw transfer]
|
493_B_2(1O49)
[Raw transfer]
|
772_B_2(1O4H)
[Raw transfer]
|
HPS_C_3(1O4O)
[Raw transfer]
|
262_B_2(1O4D)
[Raw transfer]
|
CIT_B_2(1O4L)
[Raw transfer]
|
687_B_2(1O45)
[Raw transfer]
|
256_B_2(1O4Q)
[Raw transfer]
|
1C5_A_2(1BKM)
[Raw transfer]
|
OXD_B_2(1O4N)
[Raw transfer]
|
853_A_2(1O48)
[Raw transfer]
|
299_B_2(1O4E)
[Raw transfer]
|
MLA_B_2(1O4M)
[Raw transfer]
|
903_B_2(1O46)
[Raw transfer]
|
PSN_B_2(1O4K)
[Raw transfer]
|
821_B_2(1O43)
[Raw transfer]
|
219_B_2(1O4I)
[Raw transfer]
|
I59_A_2(1O4G)
[Raw transfer]
|
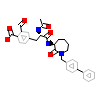
197_B_2(1O4A)
[Raw transfer]
|
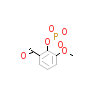
300_B_2(1O41)
[Raw transfer]
|
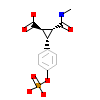
PTC_C_5(1IS0)
SRC_RSVSA
[Raw transfer]
|
PTC_D_6(1IS0)
[Raw transfer]

|
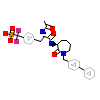
822_B_2(1O47)
[Raw transfer]
|
IS2_A_2(1O4J)
[Raw transfer]
|
790_B_2(1O4F)
[Raw transfer]
|
ACE_C_5(1A08)
SRC_HUMAN
[Raw transfer]

|
ACE_C_5(1A1E)
SRC_HUMAN
[Raw transfer]

|
ACE_C_5(1A07)
SRC_HUMAN
[Raw transfer]

|
ACE_D_6(1A09)
[Raw transfer]

|
ACE_D_6(1A08)
[Raw transfer]

|
ACE_D_6(1A1A)
[Raw transfer]

|
ACE_C_5(1A09)
SRC_HUMAN
[Raw transfer]

|
ACE_D_6(1A1B)
[Raw transfer]

|
ACE_D_6(1A1E)
[Raw transfer]

|
ACE_C_5(1A1B)
SRC_HUMAN
[Raw transfer]

|
ACE_C_5(1A1C)
SRC_HUMAN
[Raw transfer]

|
ACE_D_6(1A07)
[Raw transfer]

|
ACE_C_5(1A1A)
SRC_HUMAN
[Raw transfer]

|