QIG_A_2(3GXL)
TGFR1_HUMAN
[Raw transfer]

|
855_A_2(3HMM)
TGFR1_HUMAN
[Raw transfer]

|
ZZG_A_2(2WOT)
TGFR1_HUMAN
[Raw transfer]

|
3WO_A_2(4X2K)
TGFR1_HUMAN
[Raw transfer]

|
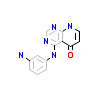
3WN_A_2(4X2J)
TGFR1_HUMAN
[Raw transfer]
|
ZZF_A_3(2WOU)
TGFR1_HUMAN
[Raw transfer]

|
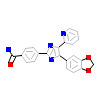
085_A_2(3TZM)
TGFR1_HUMAN
[Raw transfer]
|
J2V_A_2(5QIL)
TGFR1_HUMAN
[Raw transfer]

|
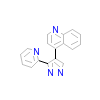
PY1_C_3(1PY5)
TGFR1_HUMAN
[Raw transfer]
|
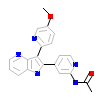
J2Y_A_2(5QIM)
TGFR1_HUMAN
[Raw transfer]
|
STU_A_2(5E8W)
TGFR1_HUMAN
[Raw transfer]

|
580_B_2(1RW8)
TGFR1_HUMAN
[Raw transfer]
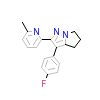
|
D0A_A_2(6B8Y)
TGFR1_HUMAN
[Raw transfer]

|
GOL_A_3(5E8T)
TGFR1_HUMAN
[Raw transfer]

|
ZUQ_A_4(5FRI)
TGFR1_HUMAN
[Raw transfer]
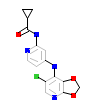
|
J2M_A_2(5QIK)
TGFR1_HUMAN
[Raw transfer]
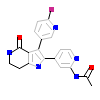
|
STU_A_2(5E8X)
TGFR1_HUMAN
[Raw transfer]

|
8LY_A_2(5USQ)
TGFR1_HUMAN
[Raw transfer]
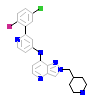
|
5L4_A_2(5E8Z)
TGFR1_HUMAN
[Raw transfer]
|
3WJ_A_2(4X2F)
TGFR1_HUMAN
[Raw transfer]

|
5L4_A_2(5E90)
TGFR1_HUMAN
[Raw transfer]

|
3WA_A_2(4X0M)
TGFR1_HUMAN
[Raw transfer]
|
3WK_A_2(4X2G)
TGFR1_HUMAN
[Raw transfer]
|