2QK_B_5(4O6L)
[Raw transfer]
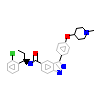
|
8Q5_A_2(5N84)
[Raw transfer]
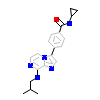
|
N66_A_2(5N87)
[Raw transfer]
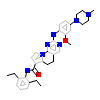
|
052_A_10(4ZEG)
[Raw transfer]
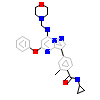
|
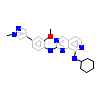
5O1_A_2(5EHO)
[Raw transfer]
|
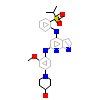
S22_A_2(3GFW)
[Raw transfer]
|
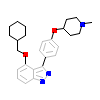
1PF_A_2(4JS8)
[Raw transfer]
|
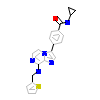
O23_A_3(3WZK)
[Raw transfer]
|
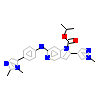
PWU_A_2(5AP6)
[Raw transfer]
|
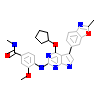
CQ7_A_2(6B4W)
[Raw transfer]
|
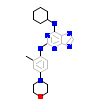
8PT_A_2(5N7V)
[Raw transfer]
|
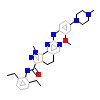
C5N_A_2(5O91)
[Raw transfer]
|
PWU_A_2(5AP2)
[Raw transfer]

|
1O5_A_2(3W1F)
[Raw transfer]
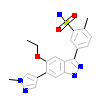
|
7RO_A_2(5AP5)
[Raw transfer]
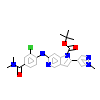
|
W2K_A_2(4CV9)
[Raw transfer]
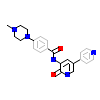
|
O38_A_3(3WYX)
[Raw transfer]
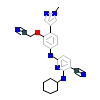
|
O43_A_2(3WZJ)
[Raw transfer]
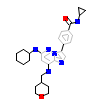
|
AD5_A_2(5LJJ)
[Raw transfer]
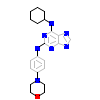
|
8QW_A_2(5N9S)
[Raw transfer]
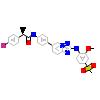
|
DYK_A_2(4D2S)
[Raw transfer]

|
5NW_A_2(5EH0)
[Raw transfer]
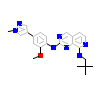
|
STU_A_3(3HMO)
[Raw transfer]

|
WBI_A_2(4CVA)
[Raw transfer]
|
8RH_A_2(5NAD)
[Raw transfer]
|
SVE_A_2(5NTT)
[Raw transfer]
|
SVE_A_2(2X9E)
[Raw transfer]

|
GOL_A_7(6GVJ)
?
[Raw transfer]

|
FMW_A_2(6H3K)
[Raw transfer]
|
O17_A_2(3WYY)
[Raw transfer]
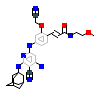
|
O22_A_10(3VQU)
[Raw transfer]
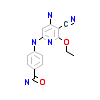
|
0SQ_A_2(5EHL)
[Raw transfer]
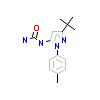
|
ATP_A_2(3HMN)
[Raw transfer]

|
7PE_A_2(3CEK)
[Raw transfer]

|
7PE_A_8(5EHY)
[Raw transfer]

|
1KA_A_3(5AP5)
[Raw transfer]
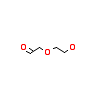
|