C0R_C_3(2A3I)
[Raw transfer]
|
EYN_A_4(6GGG)
?
[Raw transfer]
|
CV7_A_6(4UDB)
[Raw transfer]
|
1CA_D_4(1Y9R)
[Raw transfer]
|
1CA_E_5(2ABI)
MCR_HUMAN
[Raw transfer]

|
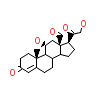
AS4_D_4(2AA2)
[Raw transfer]
|
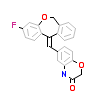
60R_A_4(5HCV)
MCR_HUMAN
[Raw transfer]
|
1CA_C_3(1Y9R)
MCR_HUMAN
[Raw transfer]

|
1CA_C_3(2AA7)
[Raw transfer]

|
1CA_D_4(2ABI)
MCR_HUMAN
[Raw transfer]

|
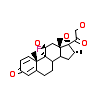
DEX_A_4(4UDA)
[Raw transfer]
|
AS4_A_4(2Q1H)
?
[Raw transfer]

|
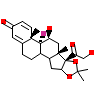
1TA_B_6(5UFS)
?
[Raw transfer]
|
1CA_F_6(2ABI)
[Raw transfer]

|
1TA_A_5(5UFS)
?
[Raw transfer]

|
DEX_A_5(3GN8)
?
[Raw transfer]

|
1CA_A_4(3RY9)
?
[Raw transfer]

|
SNL_A_4(2AB2)
MCR_HUMAN
[Raw transfer]
|
1CA_B_7(3RY9)
?
[Raw transfer]

|
EY8_A_4(6GG8)
?
[Raw transfer]
|
WFG_A_2(3WFG)
[Raw transfer]
|
LD1_A_2(3VHV)
[Raw transfer]
|
STR_B_4(2AA6)
[Raw transfer]
|
DEX_B_6(3GN8)
?
[Raw transfer]

|
SNL_A_2(3VHU)
[Raw transfer]

|
HFN_A_2(4PF3)
[Raw transfer]
|
STR_A_3(2AA6)
MCR_HUMAN
[Raw transfer]

|
STR_F_6(1YA3)
[Raw transfer]

|
STR_D_4(1YA3)
MCR_HUMAN
[Raw transfer]

|
STR_A_3(2AA5)
MCR_HUMAN
[Raw transfer]

|
STR_B_4(2AA5)
[Raw transfer]

|
WFF_A_2(3WFF)
[Raw transfer]
|
6Q0_A_8(5L7E)
[Raw transfer]
|
STR_B_9(4FN9)
?
[Raw transfer]

|
6QE_A_4(5L7G)
[Raw transfer]
|
STR_A_3(4FN9)
?
[Raw transfer]

|
SNL_B_5(2AB2)
[Raw transfer]

|
MOF_A_6(4E2J)
?
[Raw transfer]
|
STR_E_5(1YA3)
MCR_HUMAN
[Raw transfer]

|
MOF_B_7(4E2J)
?
[Raw transfer]

|
LD2_A_3(3VHV)
[Raw transfer]
|
486_A_4(4LTW)
?
[Raw transfer]
|