17H_A_3(2AMB)
[Raw transfer]
|
ENM_A_3(2PNU)
[Raw transfer]
|
TES_A_3(2AM9)
[Raw transfer]
|
TES_A_2(2YLO)
?
[Raw transfer]

|
B66_A_2(3B66)
[Raw transfer]
|
HFT_B_2(2AX6)
[Raw transfer]
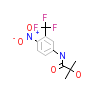
|
RLL_A_2(3RLL)
[Raw transfer]
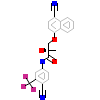
|
DHT_A_3(1T79)
[Raw transfer]

|
DHT_A_2(1T7T)
[Raw transfer]

|
R18_B_2(1XQ3)
?
[Raw transfer]
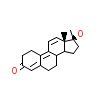
|
TES_A_2(4HLW)
[Raw transfer]

|
FHM_B_2(2AXA)
[Raw transfer]
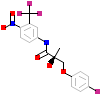
|
DHT_A_3(2AMA)
[Raw transfer]

|
R18_C_3(1XOW)
[Raw transfer]

|
DHT_A_3(1T7F)
[Raw transfer]

|
198_C_3(1Z95)
[Raw transfer]
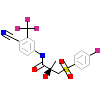
|
FHM_B_2(2AX7)
[Raw transfer]

|
TES_A_3(2YHD)
[Raw transfer]

|
DHT_C_3(1T65)
[Raw transfer]

|
TES_A_4(2Q7J)
[Raw transfer]

|
DHT_A_3(1T7R)
[Raw transfer]

|
TES_A_2(3ZQT)
[Raw transfer]

|
FHM_B_2(2AX8)
[Raw transfer]

|
DHT_A_3(3L3Z)
[Raw transfer]

|
DHT_A_4(1T74)
[Raw transfer]

|
B68_A_2(3B68)
[Raw transfer]
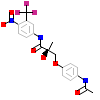
|
TES_A_3(2YLQ)
[Raw transfer]

|
DHT_C_3(1XJ7)
[Raw transfer]

|
DHT_A_13(5JJM)
[Raw transfer]

|
DHT_B_2(1I38)
[Raw transfer]

|
DHT_A_4(1T73)
[Raw transfer]

|
DHT_A_3(2PIV)
[Raw transfer]

|
DHT_A_3(2PKL)
[Raw transfer]

|
DHT_A_5(1T76)
[Raw transfer]

|
RLJ_A_2(3RLJ)
?
[Raw transfer]
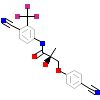
|
DHT_A_2(2PIX)
[Raw transfer]

|
DHT_C_3(1T5Z)
[Raw transfer]

|
DHT_A_3(2PIO)
[Raw transfer]

|
DHT_A_3(2Z4J)
[Raw transfer]

|
DHT_B_2(1I37)
[Raw transfer]

|
198_A_4(4OKX)
[Raw transfer]

|
DHT_A_3(2QPY)
[Raw transfer]

|
DHT_C_3(1T63)
[Raw transfer]

|
TES_A_3(2Q7L)
[Raw transfer]

|
DHT_B_19(5JJM)
?
[Raw transfer]

|
DHT_A_3(3L3X)
[Raw transfer]

|
JAD_A_2(4QL8)
[Raw transfer]
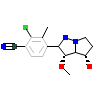
|
DHT_C_26(5JJM)
?
[Raw transfer]

|
DHT_A_4(1T7M)
[Raw transfer]

|
DHT_A_3(2PIT)
[Raw transfer]

|
51Y_A_3(5CJ6)
[Raw transfer]
|
198_A_2(4OJB)
[Raw transfer]

|
R18_C_3(2AO6)
[Raw transfer]

|
DHT_A_3(2PIR)
[Raw transfer]

|
DHT_A_3(4OFR)
[Raw transfer]

|
R18_A_2(1E3G)
[Raw transfer]

|
198_A_3(4OKT)
[Raw transfer]

|
BHM_A_2(2AX9)
[Raw transfer]
|
B67_A_2(3B67)
[Raw transfer]
|
B5R_A_2(3B5R)
[Raw transfer]
|
198_A_4(4OKB)
[Raw transfer]

|
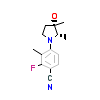
77T_A_3(5T8J)
[Raw transfer]
|
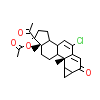
CA4_A_2(2OZ7)
[Raw transfer]
|
198_A_3(4OK1)
[Raw transfer]

|
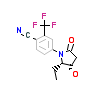
97A_A_5(5V8Q)
[Raw transfer]
|
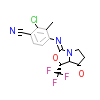
LGB_A_2(3G0W)
[Raw transfer]
|
HFT_A_3(4OJ9)
[Raw transfer]

|
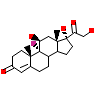
ZK5_A_2(1GS4)
[Raw transfer]
|
198_A_3(4OLM)
[Raw transfer]

|
198_A_3(4OKW)
[Raw transfer]

|
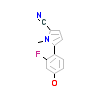
9FG_A_2(5VO4)
[Raw transfer]
|
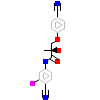
3B6_A_2(3B65)
[Raw transfer]
|
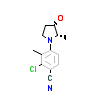
77U_A_4(5T8E)
[Raw transfer]
|
8NH_A_2(2NW4)
[Raw transfer]

|
STR_A_3(4FN9)
?
[Raw transfer]
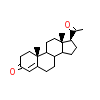
|
LG7_B_2(2IHQ)
[Raw transfer]
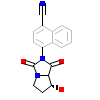
|
PK0_A_3(3V49)
[Raw transfer]
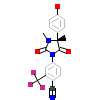
|
STR_B_9(4FN9)
?
[Raw transfer]

|
LGD_A_2(2HVC)
[Raw transfer]
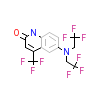
|
PK1_A_3(3V4A)
[Raw transfer]
|
RB1_A_4(2PIQ)
?
[Raw transfer]
|
K10_L_7(2PIP)
?
[Raw transfer]
|
486_A_3(4LTW)
?
[Raw transfer]
|
056_A_5(2YLP)
[Raw transfer]
|
T3_A_3(2PIW)
[Raw transfer]
|
4HY_A_3(2PIU)
[Raw transfer]
|