OHT_A_2(2GPU)
[Raw transfer]
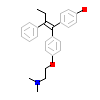
|
TXF_C_8(2EWP)
ERR3_HUMAN
[Raw transfer]
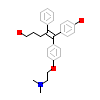
|
TXF_E_10(2EWP)
[Raw transfer]

|
1OH_A_2(2ZAS)
ERR3_HUMAN
[Raw transfer]
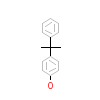
|
OHT_D_4(1VJB)
ERR3_HUMAN
[Raw transfer]

|
OHT_C_3(1VJB)
ERR3_HUMAN
[Raw transfer]

|
90C_A_4(5YSO)
ERR3_HUMAN
[Raw transfer]
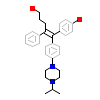
|
OHT_A_2(2P7Z)
ERR3_HUMAN
[Raw transfer]

|
TXF_A_6(2EWP)
ERR3_HUMAN
[Raw transfer]

|
90C_B_5(5YSO)
ERR3_HUMAN
[Raw transfer]

|
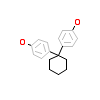
BPZ_A_2(2ZKC)
ERR3_HUMAN
[Raw transfer]
|
TXF_B_7(2EWP)
ERR3_HUMAN
[Raw transfer]

|
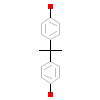
2OH_A_3(2P7G)
ERR3_HUMAN
[Raw transfer]
|
2OH_A_2(2E2R)
ERR3_HUMAN
[Raw transfer]

|
TXF_D_9(2EWP)
ERR3_HUMAN
[Raw transfer]

|
90C_C_6(5YSO)
[Raw transfer]

|
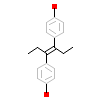
DES_H_8(1S9P)
[Raw transfer]
|
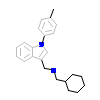
047_A_3(2PJL)
ERR1_HUMAN
[Raw transfer]
|
047_B_4(2PJL)
ERR1_HUMAN
[Raw transfer]

|
43M_A_2(2P7A)
ERR3_HUMAN
[Raw transfer]
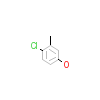
|
DES_E_5(1S9P)
ERR3_MOUSE
[Raw transfer]

|
1BA_A_5(2GPP)
ERR3_HUMAN
[Raw transfer]
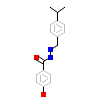
|
DES_G_7(1S9P)
ERR3_MOUSE
[Raw transfer]

|
DES_F_6(1S9P)
ERR3_MOUSE
[Raw transfer]

|
5FB_A_2(3K6P)
ERR1_HUMAN
[Raw transfer]
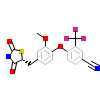
|
CHD_E_5(1S9Q)
ERR3_MOUSE
[Raw transfer]
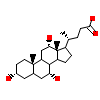
|
CHD_F_6(1S9Q)
ERR3_MOUSE
[Raw transfer]

|