A2K_B_5(4APU)
[Raw transfer]
|
AS0_B_5(4A2J)
[Raw transfer]
|
AS0_A_3(4A2J)
PRGR_HUMAN
[Raw transfer]

|
STR_A_3(1A28)
PRGR_HUMAN
[Raw transfer]
|
T98_E_5(1ZUC)
[Raw transfer]
|
R18_A_3(1E3K)
PRGR_HUMAN
[Raw transfer]
|
2S0_A_3(4OAR)
[Raw transfer]
|
R18_B_4(1E3K)
[Raw transfer]

|
STR_B_4(1A28)
[Raw transfer]

|
486_A_3(2W8Y)
PRGR_HUMAN
[Raw transfer]
|
AS4_A_4(2Q1H)
?
[Raw transfer]
|
NDR_D_4(1SQN)
[Raw transfer]
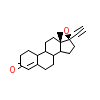
|
WOW_A_3(3KBA)
PRGR_HUMAN
[Raw transfer]
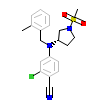
|
NOG_B_4(3D90)
[Raw transfer]
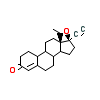
|
1CA_A_3(2Q3Y)
?
[Raw transfer]
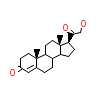
|
AS0_C_3(2OVM)
[Raw transfer]

|
T98_D_4(1ZUC)
PRGR_HUMAN
[Raw transfer]

|
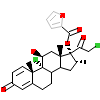
MOF_A_4(1SR7)
PRGR_HUMAN
[Raw transfer]
|
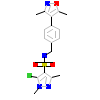
OR8_A_3(4APU)
PRGR_HUMAN
[Raw transfer]
|
AS0_A_3(2OVH)
[Raw transfer]

|
NDR_C_3(1SQN)
PRGR_HUMAN
[Raw transfer]

|
1CA_A_2(4FNE)
?
[Raw transfer]

|
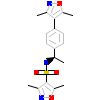
ORC_B_8(3ZRB)
[Raw transfer]
|
NOG_A_3(3D90)
PRGR_HUMAN
[Raw transfer]

|
NDR_B_5(2W8Y)
[Raw transfer]

|
1CA_B_7(3RY9)
?
[Raw transfer]

|
1CA_A_4(3RY9)
?
[Raw transfer]

|
STR_A_3(4FN9)
?
[Raw transfer]

|
WOW_B_5(3KBA)
[Raw transfer]

|
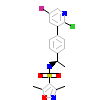
ORB_A_3(3ZRA)
PRGR_HUMAN
[Raw transfer]
|
MOF_B_5(1SR7)
[Raw transfer]

|
OR8_A_3(3ZR7)
PRGR_HUMAN
[Raw transfer]

|
OR8_B_10(3ZR7)
[Raw transfer]

|
30X_B_4(3G8O)
[Raw transfer]
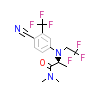
|
30X_A_3(3G8O)
PRGR_HUMAN
[Raw transfer]

|
GKK_B_9(3HQ5)
[Raw transfer]
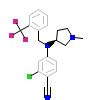
|
GKK_A_7(3HQ5)
PRGR_HUMAN
[Raw transfer]

|
OR8_A_3(3ZRB)
PRGR_HUMAN
[Raw transfer]

|
ORB_B_5(3ZRA)
[Raw transfer]

|
EDO_A_9(3G97)
GCR_RAT
[Raw transfer]

|
486_A_4(4LTW)
?
[Raw transfer]

|
EDO_A_8(3G9M)
GCR_RAT
[Raw transfer]

|
EDO_A_9(3G97)
[Raw transfer]

|
EDO_A_8(3G9M)
[Raw transfer]

|
CPS_B_11(4FN9)
?
[Raw transfer]
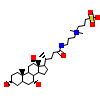
|