EPH_B_5(4DOR)
[Raw transfer]

|
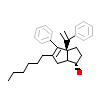
71W_A_3(5SYZ)
[Raw transfer]
|
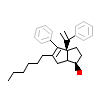
RJW_A_3(5UNJ)
[Raw transfer]
|
EPH_A_9(4PLE)
NR5A2_HUMAN
[Raw transfer]

|
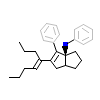
470_A_5(3PLZ)
NR5A2_HUMAN
[Raw transfer]
|
N27_A_3(6OR1)
[Raw transfer]

|
N2J_A_3(6OQY)
[Raw transfer]

|
RJW_A_3(5L11)
[Raw transfer]

|
QY4_A_3(6VIF)
[Raw transfer]

|
N1V_A_3(6OQX)
[Raw transfer]

|
EPH_F_6(1YUC)
[Raw transfer]

|
EPH_G_22(4PLE)
[Raw transfer]

|
EPH_E_16(4PLE)
NR5A2_HUMAN
[Raw transfer]

|
EPH_C_13(4PLE)
NR5A2_HUMAN
[Raw transfer]

|
EPH_A_5(4ONI)
NR5A2_HUMAN
[Raw transfer]

|
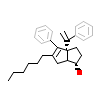
QU7_A_3(6VC2)
[Raw transfer]
|
470_B_7(3PLZ)
[Raw transfer]

|
P3A_D_4(1ZDU)
[Raw transfer]

|
P6L_B_9(4ONI)
[Raw transfer]

|
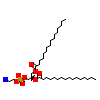
PEF_C_3(1YP0)
STF1_MOUSE
[Raw transfer]
|
P6L_B_3(3TX7)
NR5A2_HUMAN
[Raw transfer]

|
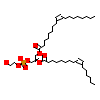
P0E_C_3(1YOW)
STF1_HUMAN
[Raw transfer]
|
PEF_E_5(1ZDT)
STF1_HUMAN
[Raw transfer]

|
PIZ_A_3(4RWV)
[Raw transfer]
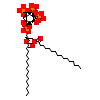
|
P6L_D_4(1YOK)
[Raw transfer]

|
PIK_A_3(4QK4)
STF1_HUMAN
[Raw transfer]

|
PEF_F_6(1ZDT)
STF1_HUMAN
[Raw transfer]

|
PIZ_A_3(4QJR)
STF1_HUMAN
[Raw transfer]

|
PLC_A_6(4DOS)
[Raw transfer]
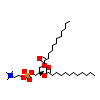
|