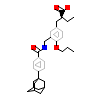
S44_A_2(2ZNN)
PPARA_HUMAN
[Raw transfer]
|
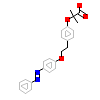
Y1N_A_2(4CI4)
PPARA_HUMAN
[Raw transfer]
|
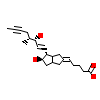
IL2_A_3(3SP6)
PPARA_HUMAN
[Raw transfer]
|
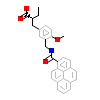
13M_A_2(3VI8)
PPARA_HUMAN
[Raw transfer]
|
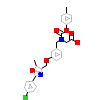
NKS_A_3(3KDU)
PPARA_HUMAN
[Raw transfer]
|
NKS_B_4(3KDU)
PPARA_HUMAN
[Raw transfer]

|
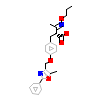
MMB_A_5(2NPA)
PPARA_HUMAN
[Raw transfer]
|
MMB_C_6(2NPA)
PPARA_HUMAN
[Raw transfer]

|
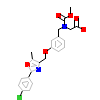
7HA_B_4(3KDT)
PPARA_HUMAN
[Raw transfer]
|
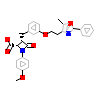
REW_A_3(2REW)
PPARA_HUMAN
[Raw transfer]
|
735_C_3(2P54)
PPARA_HUMAN
[Raw transfer]
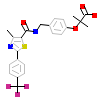
|
RO7_A_3(3G8I)
PPARA_HUMAN
[Raw transfer]
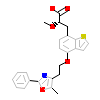
|
7HA_A_3(3KDT)
PPARA_HUMAN
[Raw transfer]

|
4M5_A_4(5AZT)
PPARA_HUMAN
[Raw transfer]
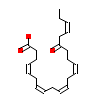
|
AZ2_A_3(1I7G)
PPARA_HUMAN
[Raw transfer]
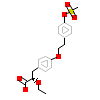
|
CTM_A_3(3FEI)
PPARA_HUMAN
[Raw transfer]
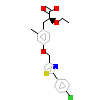
|
ET1_B_6(3ET1)
PPARA_HUMAN
[Raw transfer]
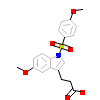
|
ET1_A_5(3ET1)
PPARA_HUMAN
[Raw transfer]

|
471_L_12(1KKQ)
PPARA_HUMAN
[Raw transfer]
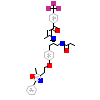
|
471_K_11(1KKQ)
PPARA_HUMAN
[Raw transfer]

|
4M5_B_5(5AZT)
PPARA_HUMAN
[Raw transfer]

|
471_I_9(1KKQ)
PPARA_HUMAN
[Raw transfer]

|
65W_A_2(5HYK)
PPARA_HUMAN
[Raw transfer]
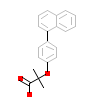
|
471_J_10(1KKQ)
PPARA_HUMAN
[Raw transfer]

|
WY1_A_5(4BCR)
PPARA_HUMAN
[Raw transfer]
|
WY1_B_8(4BCR)
PPARA_HUMAN
[Raw transfer]

|