40U_A_5(4S0T)
NR1I2_HUMAN
[Raw transfer]
|
40U_B_6(4S0T)
NR1I2_HUMAN
[Raw transfer]

|
40U_A_3(4XHD)
NR1I2_HUMAN
[Raw transfer]

|
2Q4_A_2(4NY9)
NR1I2_HUMAN
[Raw transfer]
|
SRL_A_3(3HVL)
NCOA1_HUMAN
[Raw transfer]
|
SRL_A_5(1NRL)
NR1I2_HUMAN
[Raw transfer]

|
SRL_B_6(1NRL)
NR1I2_HUMAN
[Raw transfer]

|
SRL_B_4(3HVL)
NR1I2_HUMAN
[Raw transfer]

|
444_F_6(2O9I)
NR1I2_HUMAN
[Raw transfer]
|
444_E_5(2O9I)
NR1I2_HUMAN
[Raw transfer]

|
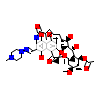
RFP_B_2(1SKX)
NR1I2_HUMAN
[Raw transfer]
|
SRL_A_2(1ILH)
NR1I2_HUMAN
[Raw transfer]

|
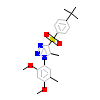
4WH_B_6(5X0R)
NR1I2_HUMAN
[Raw transfer]
|
SRL_B_6(4J5X)
NCOA1_HUMAN
[Raw transfer]

|
4WH_A_5(5X0R)
NR1I2_HUMAN
[Raw transfer]

|
SRL_A_5(4J5X)
NCOA1_HUMAN
[Raw transfer]

|
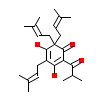
CDZ_A_2(2QNV)
NR1I2_HUMAN
[Raw transfer]
|
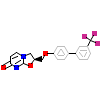
HCJ_A_3(6DUP)
NR1I2_HUMAN
[Raw transfer]
|
XGH_B_4(6BNS)
NR1I2_HUMAN
[Raw transfer]
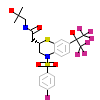
|
D7E_B_6(5A86)
NR1I2_HUMAN
[Raw transfer]
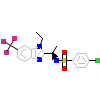
|
3WF_A_2(4X1F)
NR1I2_HUMAN
[Raw transfer]
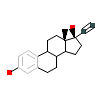
|
D7E_A_5(5A86)
NR1I2_HUMAN
[Raw transfer]

|
PNU_A_2(3R8D)
NR1I2_HUMAN
[Raw transfer]
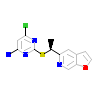
|
XGH_A_3(6BNS)
NCOA1_HUMAN
[Raw transfer]

|
3WG_A_3(4X1G)
NR1I2_HUMAN
[Raw transfer]
|