C3S_B_2(1S0X)
[Raw transfer]
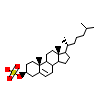
|
4D8_B_7(4S15)
[Raw transfer]
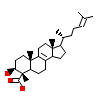
|
CLR_B_2(1N83)
[Raw transfer]
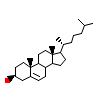
|
4D8_A_5(4S15)
RORA_HUMAN
[Raw transfer]

|
ARL_A_3(1NQ7)
RORB_RAT
[Raw transfer]
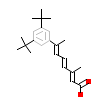
|
EF5_A_5(6G05)
RORG_HUMAN
[Raw transfer]
|
98N_A_9(5NTW)
[Raw transfer]
|
REA_C_3(1N4H)
RORB_RAT
[Raw transfer]
|
C3S_A_9(5NTI)
[Raw transfer]

|
98H_A_9(5NTN)
RORG_HUMAN
[Raw transfer]
|
4D8_A_3(4S14)
[Raw transfer]

|
STE_C_3(1K4W)
RORB_RAT
[Raw transfer]
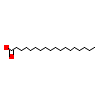
|
8YB_A_5(5NU1)
RORG_HUMAN
[Raw transfer]
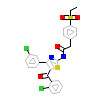
|
EEZ_A_9(6G07)
RORG_HUMAN
[Raw transfer]
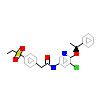
|
4LQ_A_3(5AYG)
RORG_HUMAN
[Raw transfer]
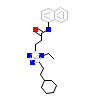
|
6Q5_A_3(5X8S)
?
[Raw transfer]
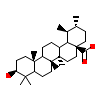
|
B6L_A_4(6J1L)
RORG_HUMAN
[Raw transfer]

|
F7M_A_3(6CN5)
RORG_HUMAN
[Raw transfer]
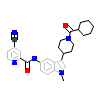
|
EE8_A_5(6FZU)
[Raw transfer]
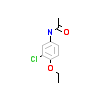
|
444_A_5(5EJV)
[Raw transfer]
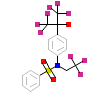
|