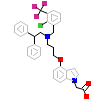
LX2_D_8(3FC6)
NR1H3_MOUSE
[Raw transfer]
|
LX2_B_6(3FC6)
NR1H3_MOUSE
[Raw transfer]

|
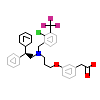
965_A_3(3IPQ)
NR1H3_HUMAN
[Raw transfer]
|
KVB_A_2(6S4T)
NR1H2_HUMAN
[Raw transfer]

|
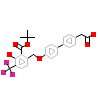
4KQ_A_3(5AVL)
NR1H3_HUMAN
[Raw transfer]
|
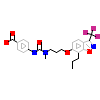
O40_B_6(3IPU)
NR1H3_HUMAN
[Raw transfer]
|
965_A_5(1PQ6)
NR1H2_HUMAN
[Raw transfer]

|
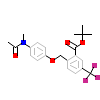
4KM_C_6(5AVI)
NR1H3_HUMAN
[Raw transfer]
|
4KM_A_5(5AVI)
NR1H3_HUMAN
[Raw transfer]

|
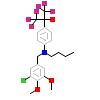
LO2_B_6(3FAL)
NR1H3_MOUSE
[Raw transfer]
|
O40_A_5(3IPU)
NR1H3_HUMAN
[Raw transfer]

|
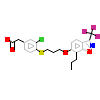
O90_A_6(3IPS)
NR1H3_HUMAN
[Raw transfer]
|
LO2_D_8(3FAL)
NR1H3_MOUSE
[Raw transfer]

|
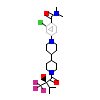
668_B_6(5HJS)
NR1H3_HUMAN
[Raw transfer]
|
668_A_5(5HJS)
NR1H3_HUMAN
[Raw transfer]

|
O90_B_7(3IPS)
NR1H3_HUMAN
[Raw transfer]

|
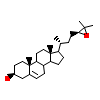
CO1_E_5(1P8D)
NR1H2_HUMAN
[Raw transfer]
|
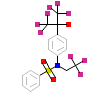
444_E_5(1UHL)
NR1H3_HUMAN
[Raw transfer]
|
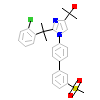
652_A_5(4RAK)
NR1H2_HUMAN
[Raw transfer]
|
KVK_A_4(6S4U)
NR1H2_HUMAN
[Raw transfer]

|
444_A_2(1UPV)
NR1H2_HUMAN
[Raw transfer]

|
444_A_2(1UPW)
NR1H2_HUMAN
[Raw transfer]

|
KUW_D_5(6S4N)
NR1H2_HUMAN
[Raw transfer]

|
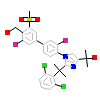
6OX_A_5(5JY3)
NR1H2_HUMAN
[Raw transfer]
|