PLM_A_3(7A78)
[Raw transfer]
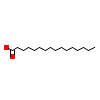
|
LG2_C_11(1H9U)
RXRB_HUMAN
[Raw transfer]
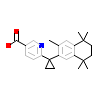
|
LG2_A_5(1H9U)
RXRB_HUMAN
[Raw transfer]

|
LG2_B_8(1H9U)
RXRB_HUMAN
[Raw transfer]

|
L79_E_5(1RDT)
RXRA_HUMAN
[Raw transfer]
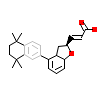
|
REA_A_11(3E00)
RXRA_HUMAN
[Raw transfer]
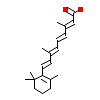
|
LG2_D_13(1H9U)
[Raw transfer]

|
MEI_F_6(1UHL)
[Raw transfer]
|
9CR_A_9(5UAN)
RXRA_HUMAN
[Raw transfer]
|
REA_B_4(3UVV)
RXRA_HUMAN
[Raw transfer]

|
REA_A_11(3DZU)
RXRA_HUMAN
[Raw transfer]

|
T9T_A_3(3KWY)
RXRA_HUMAN
[Raw transfer]
|
REA_A_5(3FC6)
RXRA_HUMAN
[Raw transfer]

|
TBY_A_3(3E94)
RXRA_HUMAN
[Raw transfer]
|
2QO_A_3(4OC7)
RXRA_HUMAN
[Raw transfer]
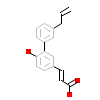
|
REA_A_5(3FAL)
RXRA_HUMAN
[Raw transfer]

|
9HF_A_6(5ZQU)
RXRA_HUMAN
[Raw transfer]
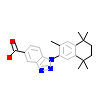
|