DEX_A_4(3MNP)
[Raw transfer]
|
DEX_A_5(3GN8)
?
[Raw transfer]

|
DEX_A_4(3MNP)
[Raw transfer]

|
CV7_A_7(4UDD)
[Raw transfer]
|
1TA_A_5(5UFS)
?
[Raw transfer]
|
DAY_A_3(3BQD)
[Raw transfer]
|
1TA_B_6(5UFS)
?
[Raw transfer]

|
1TA_A_5(5UFS)
?
[Raw transfer]

|
DEX_A_4(3MNO)
[Raw transfer]

|
DEX_B_6(3GN8)
?
[Raw transfer]

|
8W5_A_6(5NFP)
[Raw transfer]
|
DEX_A_4(4UDC)
[Raw transfer]

|
DEX_I_9(1M2Z)
[Raw transfer]

|
DEX_H_8(1M2Z)
GCR_HUMAN
[Raw transfer]

|
DEX_A_4(3MNE)
[Raw transfer]

|
NN7_A_4(4CSJ)
[Raw transfer]
|
MOF_A_3(4P6W)
[Raw transfer]
|
C0R_C_3(2A3I)
[Raw transfer]
|
B9W_A_4(6EL9)
[Raw transfer]
|
DEX_L_12(1P93)
[Raw transfer]

|
GW6_B_6(3CLD)
[Raw transfer]
|
JZN_A_7(3K23)
GCR_HUMAN
[Raw transfer]
|
DEX_J_10(1P93)
GCR_HUMAN
[Raw transfer]

|
MOF_A_6(4E2J)
?
[Raw transfer]

|
AS4_A_4(2Q1H)
?
[Raw transfer]
|
DEX_K_11(1P93)
GCR_HUMAN
[Raw transfer]

|
AS4_A_4(2Q1H)
?
[Raw transfer]

|
486_B_8(3H52)
GCR_HUMAN
[Raw transfer]
|
MOF_B_7(4E2J)
?
[Raw transfer]

|
LSJ_A_3(4LSJ)
[Raw transfer]
|
DEX_I_9(1P93)
GCR_HUMAN
[Raw transfer]

|
GW6_A_5(3CLD)
GCR_HUMAN
[Raw transfer]

|
B9Q_A_4(6EL6)
[Raw transfer]
|
LD1_A_2(3VHV)
[Raw transfer]
|
JZN_B_8(3K23)
GCR_HUMAN
[Raw transfer]

|
1CA_A_4(3RY9)
?
[Raw transfer]
|
R8C_A_4(5G5W)
[Raw transfer]
|
1CA_B_7(3RY9)
?
[Raw transfer]

|
STR_A_3(4FN9)
?
[Raw transfer]
|
486_B_13(5UC3)
[Raw transfer]

|
STR_B_9(4FN9)
?
[Raw transfer]

|
8W8_A_6(5NFT)
[Raw transfer]
|
486_A_3(5UC3)
GCR_HUMAN
[Raw transfer]

|
JZS_A_5(3K22)
GCR_HUMAN
[Raw transfer]
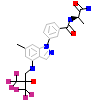
|
486_B_2(1NHZ)
[Raw transfer]

|
866_B_7(3E7C)
[Raw transfer]
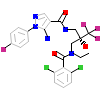
|
HJ4_B_4(6DXK)
[Raw transfer]
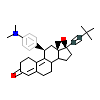
|
E7T_A_5(5G3J)
[Raw transfer]
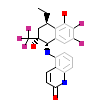
|
29M_A_5(4MDD)
GCR_HUMAN
[Raw transfer]
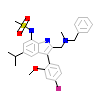
|
HJ4_A_3(6DXK)
GCR_HUMAN
[Raw transfer]

|
STR_A_2(4LTW)
?
[Raw transfer]

|
486_A_7(3H52)
GCR_HUMAN
[Raw transfer]

|
HFN_A_2(4PF3)
[Raw transfer]
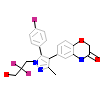
|
STR_A_2(4LTW)
?
[Raw transfer]

|
JZN_C_9(3K23)
[Raw transfer]

|
B9T_A_4(6EL7)
[Raw transfer]
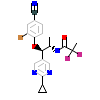
|
866_A_5(3E7C)
GCR_HUMAN
[Raw transfer]

|
DHT_B_19(5JJM)
?
[Raw transfer]

|
486_D_11(3H52)
[Raw transfer]

|
NDR_C_3(1SQN)
PRGR_HUMAN
[Raw transfer]
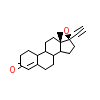
|
198_C_3(1Z95)
[Raw transfer]
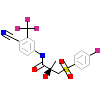
|
97A_A_5(5V8Q)
[Raw transfer]
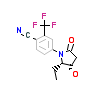
|
486_C_10(3H52)
GCR_HUMAN
[Raw transfer]

|
CPS_B_13(5UC1)
?
[Raw transfer]
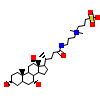
|
CPS_A_5(5UC1)
?
[Raw transfer]

|
EDO_A_3(5G5W)
[Raw transfer]

|
EDO_A_3(4CSJ)
[Raw transfer]

|