EDO_A_10(3Q4U)
ACVR1_HUMAN
[Raw transfer]

|
M2Z_B_14(6SZM)
?
[Raw transfer]

|
M2Z_A_5(6SZM)
?
[Raw transfer]

|
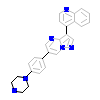
LDN_B_13(3Q4U)
ACVR1_HUMAN
[Raw transfer]
|
LU8_B_18(6SRH)
ACVR1_HUMAN
[Raw transfer]

|
IYZ_A_4(4DYM)
ACVR1_HUMAN
[Raw transfer]

|
LDN_C_20(3Q4U)
ACVR1_HUMAN
[Raw transfer]

|
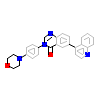
IR2_B_10(6GIN)
ACVR1_HUMAN
[Raw transfer]
|
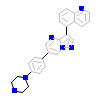
B4B_C_10(5OXG)
ACVR1_HUMAN
[Raw transfer]
|
IR2_A_3(6GIN)
ACVR1_HUMAN
[Raw transfer]

|
B4B_D_13(5OXG)
ACVR1_HUMAN
[Raw transfer]

|
MM8_A_5(6T6D)
?
[Raw transfer]

|
B4B_C_10(5OXG)
ACVR1_HUMAN
[Raw transfer]

|
LDN_A_5(3Q4U)
ACVR1_HUMAN
[Raw transfer]

|
844_A_5(4BGG)
ACVR1_HUMAN
[Raw transfer]

|
LU8_A_3(6SRH)
ACVR1_HUMAN
[Raw transfer]

|
844_B_10(4BGG)
ACVR1_HUMAN
[Raw transfer]

|
MM8_C_13(6T6D)
?
[Raw transfer]

|
LDN_A_5(3MDY)
BMR1B_HUMAN
[Raw transfer]

|
9TO_A_3(6ACR)
ACVR1_HUMAN
[Raw transfer]

|
844_C_14(4BGG)
ACVR1_HUMAN
[Raw transfer]

|
LU8_A_3(6SRH)
ACVR1_HUMAN
[Raw transfer]

|
MVE_B_12(6T8N)
?
[Raw transfer]

|
MM8_D_17(6T6D)
?
[Raw transfer]

|
MM8_B_9(6T6D)
?
[Raw transfer]

|
B4E_C_9(5OY6)
ACVR1_HUMAN
[Raw transfer]

|
GOL_A_6(4DYM)
ACVR1_HUMAN
[Raw transfer]

|
B4B_A_5(5OXG)
ACVR1_HUMAN
[Raw transfer]

|
507_A_4(3OOM)
ACVR1_HUMAN
[Raw transfer]

|
B4E_D_10(5OY6)
ACVR1_HUMAN
[Raw transfer]

|
B4B_B_8(5OXG)
ACVR1_HUMAN
[Raw transfer]

|
B4E_B_8(5OY6)
ACVR1_HUMAN
[Raw transfer]

|
844_D_16(4BGG)
ACVR1_HUMAN
[Raw transfer]

|
MVE_A_6(6T8N)
?
[Raw transfer]

|
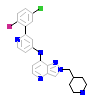
8LY_A_2(5USQ)
TGFR1_HUMAN
[Raw transfer]
|
9TO_B_7(6ACR)
ACVR1_HUMAN
[Raw transfer]

|
GOL_A_8(4DYM)
ACVR1_HUMAN
[Raw transfer]

|
EZB_A_2(6GI6)
ACVR1_HUMAN
[Raw transfer]

|
EUN_A_2(6GIP)
ACVR1_HUMAN
[Raw transfer]

|
B4E_A_5(5OY6)
ACVR1_HUMAN
[Raw transfer]

|
TAK_A_3(3Q4T)
AVR2A_HUMAN
[Raw transfer]

|
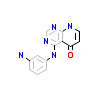
3WN_A_2(4X2J)
TGFR1_HUMAN
[Raw transfer]
|
STU_A_2(5E8X)
TGFR1_HUMAN
[Raw transfer]

|
A3F_A_17(3MTF)
ACVR1_HUMAN
[Raw transfer]

|
TAK_B_17(3Q4T)
AVR2A_HUMAN
[Raw transfer]

|
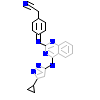
GVD_B_10(3SOC)
AVR2A_HUMAN
[Raw transfer]
|
J2V_A_2(5QIN)
TGFR2_HUMAN
[Raw transfer]

|
GVD_A_3(3SOC)
AVR2A_HUMAN
[Raw transfer]

|
STU_A_2(5E8Y)
TGFR2_HUMAN
[Raw transfer]

|
J2V_A_2(5QIN)
TGFR2_HUMAN
[Raw transfer]

|
GVD_A_3(3SOC)
AVR2A_HUMAN
[Raw transfer]

|
STU_A_2(5E8Y)
TGFR2_HUMAN
[Raw transfer]

|
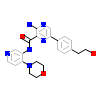
5L4_A_2(5E91)
TGFR2_HUMAN
[Raw transfer]
|
SM5_A_3(3OMV)
RAF1_HUMAN
[Raw transfer]

|
STU_A_3(3PPZ)
CTR1_ARATH
[Raw transfer]

|
STU_A_3(3P86)
CTR1_ARATH
[Raw transfer]

|
6OJ_B_8(4ASX)
AVR2A_HUMAN
[Raw transfer]

|
6OJ_A_3(4ASX)
AVR2A_HUMAN
[Raw transfer]

|
6OJ_A_3(4ASX)
AVR2A_HUMAN
[Raw transfer]

|
ADP_B_9(3G2F)
BMPR2_HUMAN
[Raw transfer]

|
ADE_A_4(2QLU)
AVR2B_HUMAN
[Raw transfer]

|
ADP_A_3(3G2F)
BMPR2_HUMAN
[Raw transfer]

|
ANP_A_2(5E92)
TGFR2_HUMAN
[Raw transfer]

|
EDO_A_8(6GIP)
ACVR1_HUMAN
[Raw transfer]

|
EDO_A_5(6I1S)
ACVR1_HUMAN
[Raw transfer]

|
EDO_A_11(6EIX)
ACVR1_HUMAN
[Raw transfer]

|
DMS_B_8(6T8N)
?
[Raw transfer]
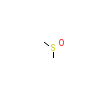
|
PG4_A_11(3H9R)
ACVR1_HUMAN
[Raw transfer]
|
DMS_B_16(6SZM)
?
[Raw transfer]
|