ZZG_A_2(2WOT)
TGFR1_HUMAN
[Raw transfer]

|
IYZ_A_4(4DYM)
ACVR1_HUMAN
[Raw transfer]

|
M2Z_B_14(6SZM)
?
[Raw transfer]

|
STU_A_2(5E8W)
TGFR1_HUMAN
[Raw transfer]

|
GOL_A_6(4DYM)
ACVR1_HUMAN
[Raw transfer]

|
JZO_E_20(3KCF)
TGFR1_HUMAN
[Raw transfer]

|
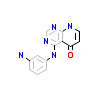
3WN_A_2(4X2J)
TGFR1_HUMAN
[Raw transfer]
|
STU_A_2(5E8X)
TGFR1_HUMAN
[Raw transfer]

|
STU_A_2(5E8X)
TGFR1_HUMAN
[Raw transfer]

|
55F_D_9(3FAA)
TGFR1_HUMAN
[Raw transfer]

|
A3F_A_2(6EIX)
ACVR1_HUMAN
[Raw transfer]

|
580_B_2(1RW8)
TGFR1_HUMAN
[Raw transfer]
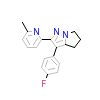
|
J2V_A_2(5QIL)
TGFR1_HUMAN
[Raw transfer]

|
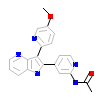
J2Y_A_2(5QIM)
TGFR1_HUMAN
[Raw transfer]
|
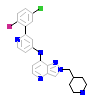
8LY_A_2(5USQ)
TGFR1_HUMAN
[Raw transfer]
|
D0A_A_2(6B8Y)
TGFR1_HUMAN
[Raw transfer]

|
3WN_A_2(4X2J)
TGFR1_HUMAN
[Raw transfer]

|
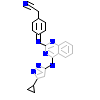
GVD_A_3(3SOC)
AVR2A_HUMAN
[Raw transfer]
|
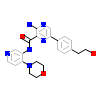
5L4_A_2(5E8Z)
TGFR1_HUMAN
[Raw transfer]
|
3WO_A_2(4X2K)
TGFR1_HUMAN
[Raw transfer]

|
8LY_A_2(5USQ)
TGFR1_HUMAN
[Raw transfer]

|
GVD_A_3(3SOC)
AVR2A_HUMAN
[Raw transfer]

|
3WJ_A_2(4X2F)
TGFR1_HUMAN
[Raw transfer]

|
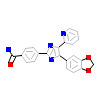
085_A_2(3TZM)
TGFR1_HUMAN
[Raw transfer]
|
ADP_B_9(3G2F)
BMPR2_HUMAN
[Raw transfer]

|
6OJ_B_8(4ASX)
AVR2A_HUMAN
[Raw transfer]

|
6OJ_A_3(4ASX)
AVR2A_HUMAN
[Raw transfer]

|
6OJ_A_3(4ASX)
AVR2A_HUMAN
[Raw transfer]

|
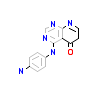
3WK_A_2(4X2G)
TGFR1_HUMAN
[Raw transfer]
|
ZUQ_A_4(5FRI)
TGFR1_HUMAN
[Raw transfer]
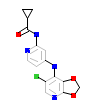
|
ADP_A_3(3G2F)
BMPR2_HUMAN
[Raw transfer]

|
ADP_A_3(3G2F)
BMPR2_HUMAN
[Raw transfer]

|
3WA_A_2(4X0M)
TGFR1_HUMAN
[Raw transfer]
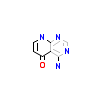
|
TRS_A_4(4X2G)
TGFR1_HUMAN
[Raw transfer]

|
EDO_A_11(6EIX)
ACVR1_HUMAN
[Raw transfer]

|
EDO_A_16(3OOM)
ACVR1_HUMAN
[Raw transfer]

|
DMS_B_16(6SZM)
?
[Raw transfer]
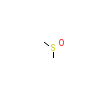
|