PEG_A_3(5M53)
NEK2_HUMAN
[Raw transfer]
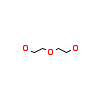
|
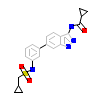
IDK_A_3(5I3R)
BMP2K_HUMAN
[Raw transfer]
|
IDK_A_3(5I3R)
BMP2K_HUMAN
[Raw transfer]

|
IDV_B_6(5I3O)
BMP2K_HUMAN
[Raw transfer]

|
IDV_A_3(5I3O)
BMP2K_HUMAN
[Raw transfer]

|
6BU_A_2(5IKW)
BMP2K_HUMAN
[Raw transfer]

|
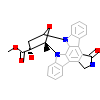
KSA_A_7(4WSQ)
AAK1_HUMAN
[Raw transfer]
|
KSA_A_7(4WSQ)
AAK1_HUMAN
[Raw transfer]

|
KSA_B_11(4WSQ)
AAK1_HUMAN
[Raw transfer]

|
3JW_A_4(4W9X)
BMP2K_HUMAN
[Raw transfer]

|
LKB_B_5(5L4Q)
AAK1_HUMAN
[Raw transfer]

|
IDK_B_5(5I3R)
BMP2K_HUMAN
[Raw transfer]

|
XIN_A_2(5TE0)
AAK1_HUMAN
[Raw transfer]

|
XIN_A_2(5TE0)
AAK1_HUMAN
[Raw transfer]

|
LKB_A_3(5L4Q)
AAK1_HUMAN
[Raw transfer]

|
YDJ_A_2(4W9W)
BMP2K_HUMAN
[Raw transfer]

|
49J_B_8(4Y8D)
GAK_HUMAN
[Raw transfer]

|
49J_A_5(4Y8D)
GAK_HUMAN
[Raw transfer]

|
YDJ_A_2(4W9W)
BMP2K_HUMAN
[Raw transfer]

|
IRE_A_3(5Y80)
GAK_HUMAN
[Raw transfer]

|
IRE_A_5(5Y7Z)
GAK_HUMAN
[Raw transfer]

|
GOL_A_14(2XNO)
NEK2_HUMAN
[Raw transfer]

|
IRE_B_8(5Y7Z)
GAK_HUMAN
[Raw transfer]

|
GGY_A_2(4AFE)
NEK2_HUMAN
[Raw transfer]

|
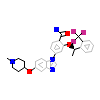
WCX_A_2(2XNP)
NEK2_HUMAN
[Raw transfer]
|
ADP_A_9(3QHW)
CDK2_HUMAN
[Raw transfer]

|
NU6_A_2(5M51)
NEK2_HUMAN
[Raw transfer]
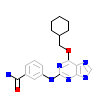
|
XK3_A_2(2XK3)
NEK2_HUMAN
[Raw transfer]
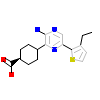
|
5Z5_A_2(2JAV)
NEK2_HUMAN
[Raw transfer]
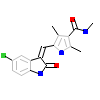
|
430_A_2(2XNN)
NEK2_HUMAN
[Raw transfer]
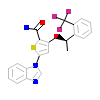
|
ATP_E_5(1UA2)
CDK7_HUMAN
[Raw transfer]

|
EDO_A_5(2W5A)
NEK2_HUMAN
[Raw transfer]

|
EDO_A_6(5M53)
NEK2_HUMAN
[Raw transfer]

|
MGF_A_12(3QHR)
CDK2_HUMAN
[Raw transfer]

|
MGF_C_26(3QHR)
CDK2_HUMAN
[Raw transfer]

|
MGF_C_18(3QHW)
CDK2_HUMAN
[Raw transfer]

|
MGF_A_12(3QHW)
CDK2_HUMAN
[Raw transfer]

|
EDO_A_5(6GVA)
CDK2_HUMAN
[Raw transfer]

|