G98_A_5(3E8D)
AKT2_HUMAN
[Raw transfer]

|
G98_B_6(3E8D)
AKT2_HUMAN
[Raw transfer]

|
X37_A_3(2XH5)
AKT2_HUMAN
[Raw transfer]

|
SMR_A_3(3QKL)
AKT1_HUMAN
[Raw transfer]

|
GVP_A_3(2UW9)
AKT2_HUMAN
[Raw transfer]

|
0XZ_A_2(4GV1)
AKT1_HUMAN
[Raw transfer]

|
X39_A_3(2X39)
AKT2_HUMAN
[Raw transfer]

|
ANP_A_3(1O6L)
AKT2_HUMAN
[Raw transfer]

|
ANP_A_3(1O6K)
AKT2_HUMAN
[Raw transfer]

|
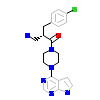
XM1_A_5(3OCB)
AKT1_HUMAN
[Raw transfer]
|
ANP_B_8(4EKK)
AKT1_HUMAN
[Raw transfer]

|
0RF_A_2(4EKL)
AKT1_HUMAN
[Raw transfer]

|
ANP_A_5(4EKK)
AKT1_HUMAN
[Raw transfer]

|
I5S_A_3(2JDO)
AKT2_HUMAN
[Raw transfer]

|
L20_A_3(2JDR)
AKT2_HUMAN
[Raw transfer]

|
XM1_B_6(3OCB)
AKT1_HUMAN
[Raw transfer]

|
WFE_A_4(3MVH)
AKT1_HUMAN
[Raw transfer]

|
SM9_A_2(3QKM)
AKT1_HUMAN
[Raw transfer]

|
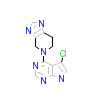
CQW_A_4(3CQW)
AKT1_HUMAN
[Raw transfer]
|
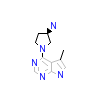
XFE_A_4(3MV5)
AKT1_HUMAN
[Raw transfer]
|
SMY_B_6(3OW4)
AKT1_HUMAN
[Raw transfer]

|
EX4_A_2(6CCY)
AKT1_HUMAN
[Raw transfer]

|
SMY_A_5(3OW4)
AKT1_HUMAN
[Raw transfer]

|
SMH_A_3(3QKK)
AKT1_HUMAN
[Raw transfer]

|
6S1_A_2(5KCV)
AKT1_HUMAN
[Raw transfer]

|
EDO_A_4(2JDO)
AKT2_HUMAN
[Raw transfer]

|