EDO_B_6(5L4Q)
AAK1_HUMAN
[Raw transfer]

|
IDV_A_3(5I3O)
BMP2K_HUMAN
[Raw transfer]

|
IDV_A_3(5I3O)
BMP2K_HUMAN
[Raw transfer]

|
6BU_A_2(5IKW)
BMP2K_HUMAN
[Raw transfer]

|
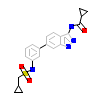
IDK_A_3(5I3R)
BMP2K_HUMAN
[Raw transfer]
|
IDV_B_6(5I3O)
BMP2K_HUMAN
[Raw transfer]

|
IDK_B_5(5I3R)
BMP2K_HUMAN
[Raw transfer]

|
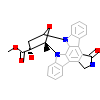
KSA_A_7(4WSQ)
AAK1_HUMAN
[Raw transfer]
|
XIN_A_2(5TE0)
AAK1_HUMAN
[Raw transfer]

|
KSA_A_7(4WSQ)
AAK1_HUMAN
[Raw transfer]

|
XIN_A_2(5TE0)
AAK1_HUMAN
[Raw transfer]

|
3JW_A_4(4W9X)
BMP2K_HUMAN
[Raw transfer]

|
KSA_B_11(4WSQ)
AAK1_HUMAN
[Raw transfer]

|
LKB_B_5(5L4Q)
AAK1_HUMAN
[Raw transfer]

|
LKB_A_3(5L4Q)
AAK1_HUMAN
[Raw transfer]

|
YDJ_A_2(4W9W)
BMP2K_HUMAN
[Raw transfer]

|
YDJ_A_2(4W9W)
BMP2K_HUMAN
[Raw transfer]

|