GOL_A_4(5A6N)
DAPK3_HUMAN
[Raw transfer]

|
ATP_B_6(2YAA)
DAPK2_MOUSE
[Raw transfer]

|
ADP_A_2(2W4K)
DAPK1_HUMAN
[Raw transfer]

|
ADP_A_2(2W4J)
DAPK1_HUMAN
[Raw transfer]

|
ACP_A_5(3ZXT)
DAPK1_HUMAN
[Raw transfer]

|
ADP_A_2(2XUU)
DAPK1_HUMAN
[Raw transfer]

|
ACP_C_12(3ZXT)
DAPK1_HUMAN
[Raw transfer]

|
ATP_A_3(2YAA)
DAPK2_MOUSE
[Raw transfer]

|
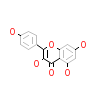
KMP_A_2(5AUX)
DAPK1_HUMAN
[Raw transfer]
|
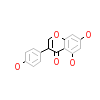
GEN_A_2(5AUZ)
DAPK1_HUMAN
[Raw transfer]
|
ACP_D_13(3ZXT)
DAPK1_HUMAN
[Raw transfer]

|
KMP_A_2(5AV2)
DAPK1_HUMAN
[Raw transfer]

|
GEN_A_2(5AV4)
DAPK1_HUMAN
[Raw transfer]

|
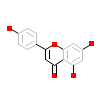
AGI_A_2(5AUV)
DAPK1_HUMAN
[Raw transfer]
|
IQU_A_5(2CKE)
DAPK2_HUMAN
[Raw transfer]

|
KMP_A_2(5AV3)
DAPK1_HUMAN
[Raw transfer]

|
IQU_D_8(2CKE)
DAPK2_HUMAN
[Raw transfer]

|
IQU_C_7(2CKE)
DAPK2_HUMAN
[Raw transfer]

|
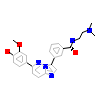
38G_A_2(4TXC)
DAPK1_HUMAN
[Raw transfer]
|
IQU_B_6(2CKE)
DAPK2_HUMAN
[Raw transfer]

|
STU_B_2(1WVY)
DAPK1_HUMAN
[Raw transfer]

|
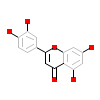
LU2_A_2(5AUU)
DAPK1_HUMAN
[Raw transfer]
|
GOL_A_9(4PF4)
DAPK1_HUMAN
[Raw transfer]

|
GOL_A_9(4PF4)
DAPK1_HUMAN
[Raw transfer]

|
2AN_A_2(5AUT)
DAPK1_HUMAN
[Raw transfer]

|
47X_A_2(5AV0)
DAPK1_HUMAN
[Raw transfer]
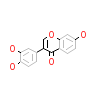
|
MRI_A_2(5AUY)
DAPK1_HUMAN
[Raw transfer]
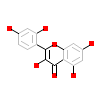
|
QUE_A_2(5AUW)
DAPK1_HUMAN
[Raw transfer]

|
DUK_A_6(5VJA)
DAPK3_HUMAN
[Raw transfer]

|
DUK_B_8(5VJA)
DAPK3_HUMAN
[Raw transfer]

|
MES_A_2(2Y0A)
DAPK1_HUMAN
[Raw transfer]
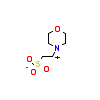
|
1PE_A_2(4B4L)
DAPK1_HUMAN
[Raw transfer]
|
DUK_C_10(5VJA)
DAPK3_HUMAN
[Raw transfer]

|
OSV_A_2(2YAK)
DAPK1_HUMAN
[Raw transfer]
|
DUK_D_13(5VJA)
DAPK3_HUMAN
[Raw transfer]

|
PGE_A_2(4UV0)
DAPK1_HUMAN
[Raw transfer]
|
PEG_A_9(6FHB)
?
[Raw transfer]
|
PEG_A_9(6FHB)
?
[Raw transfer]
|
U7E_A_3(5A6N)
DAPK3_HUMAN
[Raw transfer]

|