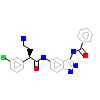
D15_A_4(2WO6)
DYR1A_HUMAN
[Raw transfer]
|
D15_B_5(2WO6)
DYR1A_HUMAN
[Raw transfer]

|
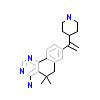
9OL_A_3(6A1G)
DYR1A_HUMAN
[Raw transfer]
|
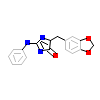
3RA_A_7(4AZE)
DYR1A_HUMAN
[Raw transfer]
|
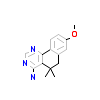
9OF_A_8(6A1F)
DYR1A_HUMAN
[Raw transfer]
|
3RA_B_8(4AZE)
DYR1A_HUMAN
[Raw transfer]

|
3RA_C_9(4AZE)
DYR1A_HUMAN
[Raw transfer]

|
9OL_B_4(6A1G)
DYR1A_HUMAN
[Raw transfer]

|
4E3_A_16(4YLL)
DYR1A_HUMAN
[Raw transfer]

|
EDO_A_12(4YLK)
DYR1A_HUMAN
[Raw transfer]

|
EDO_A_11(4YLL)
DYR1A_HUMAN
[Raw transfer]

|