AS6_A_2(2XVD)
[Raw transfer]

|
1N1_A_2(6FNM)
[Raw transfer]

|
8ZZ_A_2(5NK2)
[Raw transfer]

|
7X6_A_2(2VWZ)
[Raw transfer]

|
DWT_A_3(6FNJ)
EPHB4_HUMAN
[Raw transfer]

|
DWT_A_2(6FNG)
[Raw transfer]

|
X9F_A_2(2X9F)
[Raw transfer]

|
7X4_A_2(2VWX)
[Raw transfer]

|
32W_A_2(4BB4)
[Raw transfer]

|
8ZW_A_2(5NKI)
[Raw transfer]

|
DWT_A_2(6FNG)
[Raw transfer]

|
92Q_A_2(5NK3)
[Raw transfer]
|
8ZQ_A_2(5NKH)
[Raw transfer]

|
7O3_A_3(5MJB)
EPHB1_HUMAN
[Raw transfer]

|
90W_A_2(5NK6)
[Raw transfer]

|
DXH_A_2(6FNF)
[Raw transfer]

|
90N_A_2(5NK1)
[Raw transfer]

|
7X8_A_2(2VX1)
[Raw transfer]

|
91K_A_2(5NKD)
[Raw transfer]
|
90T_A_2(5NKC)
[Raw transfer]

|
7X5_A_2(2VWY)
[Raw transfer]

|
8ZH_A_2(5NJZ)
[Raw transfer]

|
7O3_A_3(5MJA)
EPHB1_HUMAN
[Raw transfer]

|
8ZT_A_2(5NKB)
[Raw transfer]

|
C07_A_2(4G2F)
[Raw transfer]

|
90E_A_2(5NK4)
[Raw transfer]
|
8ZN_A_2(5NKF)
[Raw transfer]
|
90Z_A_2(5NK8)
[Raw transfer]

|
DWT_B_4(6FNJ)
[Raw transfer]

|
90B_A_2(5NKE)
[Raw transfer]

|
1N1_A_2(5I9Y)
[Raw transfer]

|
GOL_A_3(2Y6M)
[Raw transfer]

|
7X7_A_2(2VX0)
[Raw transfer]

|
88Z_A_2(5IA4)
[Raw transfer]

|
8ZZ_A_2(5NK7)
[Raw transfer]

|
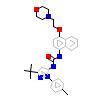
B96_A_2(4TWN)
[Raw transfer]
|
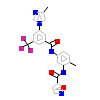
IFC_A_2(3DZQ)
[Raw transfer]
|
GV0_A_2(5IA5)
[Raw transfer]

|
IHZ_A_2(3DKO)
[Raw transfer]

|
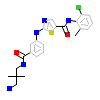
91E_A_2(5NK0)
[Raw transfer]
|
Q7M_A_2(4P5Z)
[Raw transfer]

|
GOL_A_5(3KUL)
EPHA8_HUMAN
[Raw transfer]

|
90K_A_2(5NK5)
[Raw transfer]

|
1N1_A_2(2Y6O)
[Raw transfer]

|
912_A_2(5NK9)
[Raw transfer]

|
6P8_A_2(5L6P)
[Raw transfer]

|
8ZK_A_2(5NKG)
[Raw transfer]

|
6P6_A_2(5L6O)
[Raw transfer]

|
Q9G_A_8(2XYU)
[Raw transfer]

|
DXK_A_2(6FNK)
[Raw transfer]

|
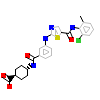
91H_A_2(5NKA)
[Raw transfer]
|
DXK_A_2(6FNK)
[Raw transfer]

|
627_A_2(5I9Z)
[Raw transfer]

|
STU_A_3(2YN8)
EPHB4_HUMAN
[Raw transfer]

|
ANP_C_3(1MQB)
EPHA2_HUMAN
[Raw transfer]

|
DXK_C_21(6FNH)
[Raw transfer]

|
ANP_D_4(1MQB)
[Raw transfer]

|
DXK_A_4(6FNH)
EPHA2_HUMAN
[Raw transfer]

|
37W_A_2(4TWO)
[Raw transfer]
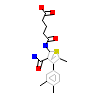
|
A5B_A_4(5IA0)
EPHA2_HUMAN
[Raw transfer]

|
30K_A_2(4AW5)
[Raw transfer]

|
L66_A_2(5IA2)
[Raw transfer]

|
STU_B_4(2YN8)
[Raw transfer]

|
A5B_B_11(5IA0)
EPHA2_HUMAN
[Raw transfer]

|
AGS_A_2(5I9V)
[Raw transfer]
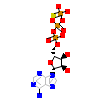
|
ANP_A_4(2QO7)
[Raw transfer]

|
DB8_A_2(5I9X)
[Raw transfer]
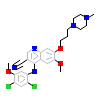
|
25Q_A_2(4P4C)
[Raw transfer]
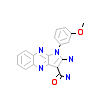
|
ZZL_A_2(5IA1)
[Raw transfer]

|
A5B_C_21(5IA0)
[Raw transfer]

|
ADP_D_12(2HEN)
[Raw transfer]

|
ADP_C_11(2HEN)
EPHB2_MOUSE
[Raw transfer]

|
DXK_B_16(6FNH)
EPHA2_HUMAN
[Raw transfer]

|
ANP_A_5(2QOC)
[Raw transfer]

|
ADP_B_10(2HEN)
EPHB2_MOUSE
[Raw transfer]

|
ANP_A_3(2QO9)
[Raw transfer]

|
ANP_A_4(3FXX)
[Raw transfer]

|
ADP_A_9(2HEN)
EPHB2_MOUSE
[Raw transfer]

|
ANP_A_3(2QOQ)
[Raw transfer]

|
ANP_A_5(2QOC)
[Raw transfer]

|
ANP_A_2(5I9W)
[Raw transfer]

|
ANP_D_4(1JPA)
[Raw transfer]

|
ANP_C_3(1JPA)
EPHB2_MOUSE
[Raw transfer]

|