99J_A_2(5VD4)
[Raw transfer]
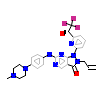
|
98D_A_2(5VDA)
[Raw transfer]

|
8X7_A_2(5V5Y)
[Raw transfer]

|
99V_A_2(5VD7)
[Raw transfer]
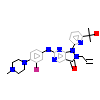
|
98G_A_2(5VD9)
[Raw transfer]

|
98M_A_2(5VD8)
[Raw transfer]

|
99M_A_2(5VD5)
[Raw transfer]

|
8X7_A_2(5VDK)
WEE2_HUMAN
[Raw transfer]

|
8X7_A_2(5VDK)
WEE2_HUMAN
[Raw transfer]

|
770_A_3(2Z2W)
[Raw transfer]

|
770_A_3(2Z2W)
[Raw transfer]

|
396_A_3(3BI6)
[Raw transfer]
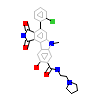
|
839_A_2(2IN6)
[Raw transfer]
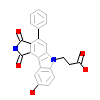
|
P91_A_3(3CQE)
[Raw transfer]

|
DB8_A_2(5VC3)
[Raw transfer]
|
P48_A_2(5VC6)
[Raw transfer]

|
34W_A_2(5VD2)
[Raw transfer]

|
XZN_A_2(5VC4)
[Raw transfer]

|
96M_A_2(5VC5)
[Raw transfer]

|