453_A_4(5HE0)
ARBK1_BOVIN
[Raw transfer]
|
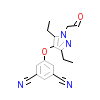
AFM_A_4(5WG3)
ARBK1_HUMAN
[Raw transfer]

|
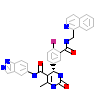
ZS2_A_4(5HE1)
ARBK1_HUMAN
[Raw transfer]
|
F0S_A_4(5HE2)
ARBK1_BOVIN
[Raw transfer]

|
EJS_A_4(6C2Y)
ARBK1_HUMAN
[Raw transfer]

|
AFV_A_4(5WG4)
ARBK1_HUMAN
[Raw transfer]

|
QRX_A_4(3PVW)
ARBK1_BOVIN
[Raw transfer]

|
T0E_A_4(5UKM)
ARBK1_BOVIN
[Raw transfer]

|
BA1_A_4(3KRW)
ARBK1_HUMAN
[Raw transfer]

|
QRW_A_2(5UUU)
ARBK1_HUMAN
[Raw transfer]

|
QRW_A_4(3PVU)
ARBK1_BOVIN
[Raw transfer]

|
BA1_A_4(3KRX)
ARBK1_HUMAN
[Raw transfer]

|
FF1_A_5(5HE3)
ARBK1_BOVIN
[Raw transfer]

|
8DJ_A_5(5UKK)
ARBK1_HUMAN
[Raw transfer]

|
KZQ_A_4(4PNK)
ARBK1_HUMAN
[Raw transfer]

|
8PV_A_2(5UVC)
ARBK1_HUMAN
[Raw transfer]

|
29X_A_5(4MK0)
ARBK1_HUMAN
[Raw transfer]

|
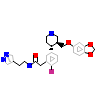
SIX_A_5(5UKL)
ARBK1_HUMAN
[Raw transfer]
|