7G6_A_2(5P9G)
[Raw transfer]

|
83P_A_2(5U9D)
[Raw transfer]

|
3OV_A_2(4RFZ)
[Raw transfer]

|
3P0_A_2(4RG0)
[Raw transfer]

|
2V2_A_2(4OTQ)
[Raw transfer]

|
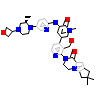
9AJ_A_2(5VFI)
[Raw transfer]
|
N42_A_5(5J87)
BTK_HUMAN
[Raw transfer]

|
GMW_A_2(6HRT)
?
[Raw transfer]

|
N42_C_7(5J87)
BTK_HUMAN
[Raw transfer]

|
2V3_A_2(4OTR)
[Raw transfer]

|
GMQ_A_2(6HRP)
?
[Raw transfer]

|
KLM_A_2(6NFH)
?
[Raw transfer]

|
3OU_A_2(4RFY)
[Raw transfer]

|
746_A_3(4Y93)
?
[Raw transfer]

|
2VL_A_2(5P9F)
[Raw transfer]

|
BNB_A_2(6EP9)
[Raw transfer]

|
N42_B_6(5J87)
BTK_HUMAN
[Raw transfer]

|
6XL_A_7(5KUP)
[Raw transfer]

|
N42_D_8(5J87)
[Raw transfer]

|
7G7_A_2(5P9H)
[Raw transfer]

|
BXM_A_4(6AUB)
[Raw transfer]

|
9B1_A_8(5VGO)
[Raw transfer]

|
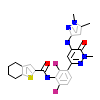
DXM_A_5(6BKW)
[Raw transfer]
|
746_A_6(3OCS)
[Raw transfer]

|
GJ7_A_2(6DI5)
[Raw transfer]

|
GJA_A_2(6DI3)
[Raw transfer]

|
5WH_A_2(5FBO)
[Raw transfer]

|
HRA_A_2(6E4F)
[Raw transfer]

|
2V1_A_2(4OT6)
[Raw transfer]

|
481_A_2(4OT5)
[Raw transfer]

|
8E8_A_2(5P9J)
[Raw transfer]

|
4C9_B_22(4YHF)
[Raw transfer]

|
1E8_A_2(5P9I)
[Raw transfer]

|
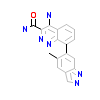
4L6_A_6(4Z3V)
[Raw transfer]
|
5WF_D_7(5FBN)
[Raw transfer]

|
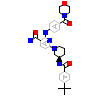
4UQ_A_3(5BPY)
BTK_HUMAN
[Raw transfer]
|
KHD_A_2(6N9P)
?
[Raw transfer]

|
DTJ_A_4(6BIK)
[Raw transfer]

|
JVP_A_3(6MNY)
?
[Raw transfer]

|
4RV_A_6(4ZLZ)
[Raw transfer]

|
DVD_A_6(6BKH)
[Raw transfer]

|
1N1_A_2(3K54)
[Raw transfer]

|
DVJ_A_5(6BKE)
[Raw transfer]

|
73T_A_2(5T18)
[Raw transfer]
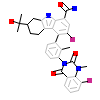
|
GJD_A_2(6DI1)
[Raw transfer]

|
BXJ_A_5(6AUA)
[Raw transfer]

|
4C9_A_3(4YHF)
BTK_HUMAN
[Raw transfer]

|
5WF_C_3(5FBN)
BTK_HUMAN
[Raw transfer]

|
027_A_2(3PIX)
[Raw transfer]

|
6MV_A_3(5JRS)
BTK_HUMAN
[Raw transfer]

|
B43_A_2(3GEN)
[Raw transfer]
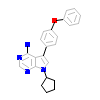
|
04L_A_2(3PJ3)
[Raw transfer]

|
KLP_A_2(6NFI)
?
[Raw transfer]

|
7G8_A_2(5P9K)
[Raw transfer]

|
3YO_A_7(4RX5)
[Raw transfer]

|
6MV_B_4(5JRS)
[Raw transfer]

|
4US_A_2(5BQ0)
[Raw transfer]

|
DY4_A_5(6BLN)
[Raw transfer]

|
7GB_A_2(5P9M)
[Raw transfer]

|
GJG_A_2(6DI0)
[Raw transfer]

|
4UQ_B_4(5BPY)
[Raw transfer]

|
4RU_A_6(4ZLY)
[Raw transfer]

|
7G9_A_2(5P9L)
[Raw transfer]

|
03C_A_2(3PIZ)
[Raw transfer]
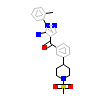
|
GJJ_A_2(6DI9)
[Raw transfer]
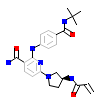
|
LHL_A_2(3PJ1)
[Raw transfer]

|
04K_A_2(3PJ2)
[Raw transfer]

|
1N1_A_2(3OCT)
[Raw transfer]

|
585_A_2(3PIY)
[Raw transfer]

|