DXV_A_2(6BKU)
KKCC2_HUMAN
[Raw transfer]

|
F6J_A_3(6CMJ)
KKCC2_HUMAN
[Raw transfer]

|
8R4_A_2(5UY6)
KKCC2_HUMAN
[Raw transfer]

|
8R7_A_2(5UYJ)
KKCC2_HUMAN
[Raw transfer]

|
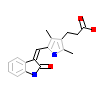
SU6_A_2(5YVC)
KKCC2_HUMAN
[Raw transfer]
|
BI9_A_2(6BQL)
KKCC2_HUMAN
[Raw transfer]

|
609_A_2(2ZV2)
KKCC2_HUMAN
[Raw transfer]

|
E5J_B_4(6BRC)
KKCC2_HUMAN
[Raw transfer]

|
6T2_A_2(6BQP)
KKCC2_HUMAN
[Raw transfer]

|
34U_A_2(6BLE)
KKCC2_HUMAN
[Raw transfer]

|
E5J_A_3(6BRC)
KKCC2_HUMAN
[Raw transfer]

|
R78_A_2(6BQQ)
KKCC2_HUMAN
[Raw transfer]

|
92C_A_2(5YVB)
KKCC2_HUMAN
[Raw transfer]

|
91L_A_2(5YV8)
KKCC2_HUMAN
[Raw transfer]

|
91X_A_2(5YVA)
KKCC2_HUMAN
[Raw transfer]

|
91O_A_2(5YV9)
KKCC2_HUMAN
[Raw transfer]

|
9JS_A_2(5VT1)
KKCC2_HUMAN
[Raw transfer]

|