K0X_A_6(5K0X)
MERTK_HUMAN
[Raw transfer]

|
CKJ_B_7(3TCP)
[Raw transfer]

|
4MH_A_6(4MHA)
MERTK_HUMAN
[Raw transfer]

|
K0X_B_9(5K0X)
[Raw transfer]

|
MH7_A_8(4MH7)
MERTK_HUMAN
[Raw transfer]

|
CKJ_A_3(3TCP)
MERTK_HUMAN
[Raw transfer]

|
6Q1_A_6(5K0K)
MERTK_HUMAN
[Raw transfer]

|
4MH_B_11(4MHA)
[Raw transfer]

|
24K_B_12(4M3Q)
[Raw transfer]

|
24K_A_8(4M3Q)
MERTK_HUMAN
[Raw transfer]

|
6Q1_B_11(5K0K)
[Raw transfer]

|
OLP_A_8(3BPR)
MERTK_HUMAN
[Raw transfer]

|
7AE_A_3(5TD2)
MERTK_HUMAN
[Raw transfer]

|
79Y_A_3(5TC0)
MERTK_HUMAN
[Raw transfer]
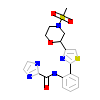
|
7YS_B_4(5U6C)
[Raw transfer]
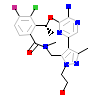
|
OLP_B_9(3BPR)
MERTK_HUMAN
[Raw transfer]

|
7YS_A_3(5U6C)
MERTK_HUMAN
[Raw transfer]

|
ANP_A_6(2P0C)
MERTK_HUMAN
[Raw transfer]

|
ANP_B_7(2P0C)
[Raw transfer]

|
OLP_D_11(3BPR)
[Raw transfer]

|
ADP_B_11(3BRB)
[Raw transfer]

|