AQ5_A_5(5OTF)
MRCKB_HUMAN
[Raw transfer]

|
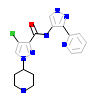
3FV_A_2(4UAL)
MRCKB_HUMAN
[Raw transfer]
|
ADP_A_2(4UAK)
MRCKB_HUMAN
[Raw transfer]

|
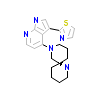
AQE_A_2(5OTE)
MRCKB_HUMAN
[Raw transfer]
|
M77_A_3(3TKU)
MRCKB_HUMAN
[Raw transfer]

|
M77_B_6(3TKU)
MRCKB_HUMAN
[Raw transfer]

|
NM7_A_3(3QFV)
MRCKB_HUMAN
[Raw transfer]

|
NM7_B_5(3QFV)
MRCKB_HUMAN
[Raw transfer]

|